
Genomic profiling of cell-free circulating tumor DNA in patients with colorectal cancer and its fidelity to the genomics of the tumor biopsy
Introduction
Liquid biopsy using comprehensive genomic profiling (CGP) is becoming a more widely used diagnostic tool for identifying genomic alterations to guide therapy and prognosis. It is largely being used in patients where tissue may not be available or is limited, where tissue-based genomic testing has failed, where the patient is too ill for an invasive biopsy, or at disease progression. Despite the continued increase in the utilization of liquid biopsy, there remains a dearth of data on the concordance, positive and negative percent agreement (PPA and NPA), and the positive and negative predictive value (PPV and NPV) of NGS liquid biopsy sequencing results with those obtained from a tissue sample obtained and sequenced at the same time. As tissue-based testing has been the “gold standard” for clinical genomic testing, it is critical to have robust data to demonstrate how well liquid biopsy recapitulates the genomics of tumors. In addition, it is important to understand the appropriate clinical settings for liquid biopsy, as well as its limitations.
Colorectal cancer (CRC) has benefited from extended KRAS and BRAF testing to inform the use of anti-EGFR antibody therapy (1). Further, other molecular predictors of efficacy of anti-EGFR antibody therapy, and genomically matched therapies such as HER2 and ALK inhibitors, as well as acquired resistance alterations, have emerged as important diagnostic and therapeutic markers required for the treatment of CRC (2-4). As such, genomic profiling has become a well-accepted tool for the classification of patients as candidates for therapy. In most cases, tumor tissue is the preferred specimen source for CGP; in some cases, tissue is not available or would not be practical to obtain-liquid biopsy, using circulating tumor DNA (ctDNA), offers a viable alternative specimen for analysis. FoundationACT™ is a hybrid capture-based genomic profiling assay for ctDNA (Foundation Medicine, Cambridge, MA, USA); the clinical utility study (CUS) (NCT02620527) is a multi-center prospective clinical study for multiple solid tumor types designed to determine whether this ctDNA assay could reliably identify alterations that were detected in paired tissue samples from the same patient taken at the same or at a later time. In the subset of the study reported here, paired liquid and tissue biopsy samples from 96 patients with CRC were analyzed by performing CGP of the tumor (using the FoundationOne® assay) and the blood plasma (using the FoundationACT™ assay). To our knowledge, this is one of the largest correlative studies of the genomic sequencing of matched tissue and liquid biopsy in CRC.
Methods
Patient population
Patients were prospectively consented and the IRB protocol was approved by Western Institutional Review Board (Protocol No. 20152817). Enrollment in the study required a previously performed FoundationOne® assay, and all paired FoundationACT™ tests were completed on blood plasma samples that were collected after the tumor sample collection and FoundationOne® testing. Liquid biopsy testing was done at the discretion of the clinician at variable time intervals after tissue sample collection (0–709 days). Clinical data were collected at the time of liquid biopsy and included stage of cancer, tissue biopsy site, metastatic sites, therapy before tumor biopsy, and therapy before liquid biopsy. Outcome data were not available. Patient demographics are included in Table 1.

Full table
Plasma and tissue CGP
Paired specimens from 96 individuals with CRC were analyzed with CGP of the solid tumor by the FoundationOne® assay and of the blood plasma sample by the FoundationACT™ assay. FoundationACT™ is the predecessor assay of current assay from Foundation Medicine: FoundationOne Liquid. FoundationACT™ detects substitutions, indels, copy number amplifications, and rearrangements in 62 genes and targets ~141 kbp of the human genome including all exons of 27 genes, selected exons of an additional 34 genes (133 exons), selected introns of 6 genes frequently involved in genomic rearrangements in cancer (12 introns), and the TERT promoter region that is recurrently mutated in cancer, as previously described (5). FoundationOne® includes all the targeted regions of the FoundationACT™ assay baitset as well as additional regions and genes. To accurately compare the tissue and liquid biopsy results, the intersection of the sequences targeted by both assays was used. For short variants and rearrangements, the analysis was done down to the coordinate level (i.e., the coordinates of the alteration in question must have been within both baitsets for it to be counted) and only known/likely alterations were included; for copy number amplifications, the comparison was done at gene level.
Because all the content of FoundationACT™ is included in FoundationOne®, all the targets of FoundationACT™ were included in the analysis. For FoundationOne®, only the content that was shared with FoundationACT™ is reported.
For genomic profiling of ctDNA from plasma, 20 mL of peripheral whole blood was collected; 20–100 ng cell-free DNA (cfDNA) was extracted from plasma and subjected to genomic profiling, with greater than 30,000× raw coverage, 5,000× unique coverage and approximately 3,000× redundant (i.e., error-corrected) coverage (6). CGP of all the solid tumor samples required >20% tumor content and was performed to a median exon coverage of ≥500× using the FoundationOne® assay as previously described (7).
All testing was performed in a CLIA-certified, CAP-accredited and New York State-accredited laboratory (Foundation Medicine, Cambridge, MA, USA).
Determination of ctDNA content in cell-free DNA fraction
Maximum somatic allele frequency (MSAF), a method of estimating the fraction of tumor-derived ctDNA versus that of total cfDNA, was calculated for all the samples. MSAF was determined by calculating the allele fraction (AF) for all known somatic, likely somatic, and variants of unknown significance (VUS) substitution alterations detected on non-PCR-duplicate read pairs. Common and rare germline variants found in the ExAC database, dbSNP v135, and 1,000 genomes database were excluded from the MSAF calculation. A calculated MSAF value of 0 indicates that no ctDNA was detected in the sample.
Statistical and data analysis
The 74 QC-passed liquid biopsy cases that had MSAF >0 were used to determine positive percent agreement (PPA) to the matched tissue biopsy genomics for all short variants. Amplifications were excluded in the analysis as they are more difficult to detect in liquid biopsy and sensitivity of detection is dependent on tumor fraction (6).
PPA and NPA, PPV and NPV, as well as total concordance between liquid and tissue biopsy were calculated for KRAS G12X; the remaining alterations in KRAS, NRAS, and BRAF were not calculated as their frequencies were very low. PPV was calculated as true positives/false positive + true positives; NPV was calculated as true negatives/true negatives + false negatives; PPA was calculated as true positives/true positive + false negatives; NPA was calculated as true negative/true negatives + false positives. Rearrangements and copy number amplifications were calculated separately.
Differences in MSAF and clinical variables were assessed by the Mann-Whitney U test for two-group comparisons. The significance of PPA difference (%) between two groups was determined using the comparison of proportions “N-1” Chi-squared test (8). All statistical tests were two-sided, and a P value of <0.05 was considered statistically significant.
Results
Patient characteristics
The patient demographic data are described in Table 1. The majority of cases were stage IV CRC (71; 74%) at tissue collection. Sixty-two (65%) of the biopsies were from metastatic sites. For all cases, tissue biopsies preceded blood collection and the median time between the tissue biopsy and liquid biopsy was 224 (range, 0–709) days. In terms of therapy, 36 (38%) of the subjects had received chemotherapy or radiation prior to the tumor resection/biopsy, and 86 (90%) of the patients had received chemotherapy or radiation therapy prior to the liquid biopsy. The average number of therapies between the tissue and liquid biopsy was 1.7 (Table 1).
Liquid biopsy quality metrics and MSAF
Ninety-two liquid biopsy cases passed all quality control (QC) metrics. One of the critical issues for accurately determining if liquid biopsy can identify the same alterations found in the matched tissue biopsy using NGS is the determination of which liquid biopsy specimens contain ctDNA. Therefore, only samples containing ctDNA were used for our analysis, while cases without detected ctDNA were excluded. MSAF was used for determination of the ctDNA fraction (9). Out of 92 cases, 18 cases (19.6%) were found to have an MSAF of 0, indicating that there was no evidence of ctDNA in the sample. It has been shown that ctDNA concentration can be affected by therapy, including the absence of or decreased ctDNA after therapy (10,11); in this cohort, 90% of the patients had received chemotherapy or radiation prior to the blood draw for the liquid biopsy. The median MSAF value for the liquid biopsy samples was 1.56% (range, 0–60.2%). MSAF was analyzed with respect to different sub-groups of the cohort. There was a statistically significant difference in MSAF between stage II/III versus stage IV cases [median 0.9% (n=24) vs. median 3.1% (n=68); P=0.02 (Mann-Whitney U test)]. A similar observation has been made previously, where fewer early stage cancer cases had detectable ctDNA than advanced cancer cases (12).
Comparison of the genomic landscape of ctDNA and tumor tissue
For the 74 liquid biopsy cases that had detectable ctDNA (MSAF >0), the liquid biopsy hybrid capture NGS sequencing identified at least one genomic alteration of any of the 4 alteration types in 68 (92%) of the cases, with an average of 2.25 genomic alterations per case. For the matched 74 tissue cases, hybrid capture NGS tissue biopsy sequencing detected at least one genomic alteration in 72 (97%) of the cases, with an average of 2.0 genomic alterations per case. In the overall set of 96 matched tissue samples, tissue biopsy sequencing detected at least one genomic alteration in 88 (92%) of the cases, with an average of 2.1 genomic alterations per case. There were 8 cases with no alterations detected in the tissue or in the corresponding liquid biopsy cases; 4 of these cases failed QC for low coverage in the liquid biopsy and 4 lacked evidence of ctDNA (MSAF =0). In all 8 cases, tumor content was >20% by H&E histology analysis.
Fidelity between tissue and liquid biopsy assay in matched liquid and tissue for the detection of indels and substitutions
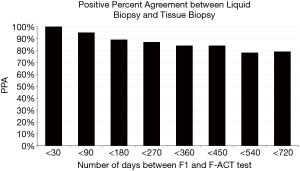
To determine how well liquid biopsy sequencing recapitulated the alterations identified in tissue biopsy sequencing, 74 matched cases that contained evidence of ctDNA (MSAF >0) were analyzed for PPA for the short variants; for KRAS G12X PPA, NPA, PPV, NPV, and overall positive agreement (OPA) was calculated. Only genes and exons covered by the tissue assay that were congruent with the same exons and genes in the liquid assay were included in the analysis. Blood samples for liquid biopsy were taken at various times after the tissue biopsy or tumor resection, and the temporal gap between the tissue and liquid biopsies varied from 0 to 709 days. We hypothesized that time between the tumor biopsy and the liquid biopsy would have an inverse relationship with the concordance between the tissue and liquid biopsy; as such, we analyzed the data across groups of increasing inclusions of cases with longer timespans between tissue and liquid biopsy (<30, <90, <180, <270, <360, <450, <540, and <720 days). PPA for the two assays in the <30 day group was 100% and in the <90 day group was 95%, whereas the overall PPA (<720 days) was 79% (Tables 2,3 and Figure 1).
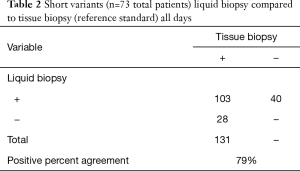
Full table
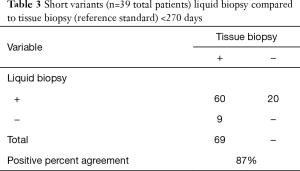
Full table
In addition, we split the dataset into two groups that represented a shorter temporal gap (<270 days; 39 cases) and a longer temporal gap (≥270 days; 34 cases). The 270 day time-point was chosen as approximately ½ the cases were collected <270 and the other ½ were collected ≥270 days; with this cutoff there are sufficient numbers of patients in each group to derive statistically significance for concordance measures. Not included in these groups was one case where no alterations were identified in either the tissue or liquid biopsy. PPA at <270 days (87%) and was significantly higher than at ≥270 days (69%) as determined by a comparison of proportions “N-1” Chi-squared test (P=0.01, 18% difference, 3.83–31.7%, 95% CI). The difference in PPA between these groups could be partially attributed to lower amounts of ctDNA in the ≥270-day group, as the median MSAFs of the two groups were notably different (6.4% in the <270-day group vs. 2.24% in the ≥270-day group); however, this difference was not statistically significant (P=0.089, Mann Whitney U test).
We also specifically analyzed the data with respect to the set of gene alterations outlined in the National Comprehensive Cancer Network (NCCN) Guidelines: KRAS, NRAS, and BRAF. We included all known/likely short variants in these genes for analysis. This analysis revealed the PPA of indels/substitutions in the NCCN group of gene alterations was 80% (Table 4), which was similar to the entire set of genes included in the liquid assay (79%). In addition, when these groups were again split into two temporally distinct groups (<270 and ≥270 days), the PPA was higher in the <270 days group (90%) as compared the ≥270 days group (68%). This difference was found to be statistically significant by a comparison of proportions “N-1” Chi-squared test (P=0.03), 22% difference, 1.79–43.30%, 95% CI). We also analyzed concordance (PPA, NPA, PPV, NPV, and OPA) between tissue and liquid for KRAS G12X for <720 and <270 days (Tables 5,6). For all cases (<720 days), the concordance was high: OPA (93%), PPA (85%), NPA (98%), PPV (96%), NPV (92%); concordance increased in the <270 day group: OPA (97%), PPA (93%), NPA (100%), PPV (100%), NPV (96%) (Table 6).
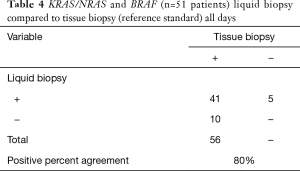
Full table
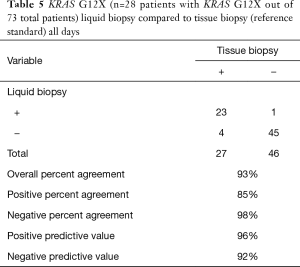
Full table
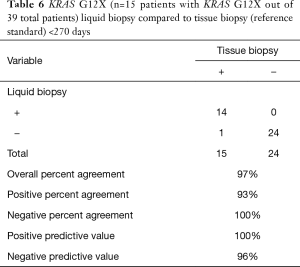
Full table
Lastly, we examined the 28 indels/substitutions that were only found in the tissue and not in the liquid biopsies. These 28 indels/substitutions were detected in 16 distinct tissue specimens. The median time between the tissue and liquid biopsy in these cases was 380 days, as compared to the median of the entire data set (224 days). There were 10 instances of KRAS alterations that are predicted to confer reduced sensitivity to EGFR inhibitors (cetuximab and panitumumab) that were only seen in the tissue. These cases were from 9 patients including 2 with stage II, 2 with stage III, and 5 with stage IV CRC, and all these patients had received chemotherapy in the intervening time between the tissue biopsy and liquid biopsy (although none had received anti-EGFR therapy). In general, the MSAF of the liquid biopsies of this 9-patient KRAS-mutated group was low, with a median MSAF value of 0.26%. We analyzed the raw sequencing data on these 10 KRAS alterations, and interestingly, there was some evidence of a KRAS alteration in 3 of these cases, but the number of unique reads with the mutation did not meet the current thresholds for reporting. The other 18 alterations found in the tissue biopsy but not the liquid biopsy included 11 TP53 alterations, 6 PIK3CA alterations, and 1 EZH2 alteration. The PI3KCA alterations were found in the two hotspots where 80% of PI3KCA alterations occur—the helicase domain of exon 9 (codon 542 and 545) and the kinase domain in exon 20 (codon 1047) (13).
The liquid biopsy identified more alterations than tissue biopsy for regions covered by both assays: 40 were unique to liquid, whereas 28 were unique to tissue (Figure 2). These 40 unique liquid biopsy alterations were from 28 patients—22 with stage IV CRC and 6 with stage III CRC. There were 2 KRAS alterations identified (G12V and Q61H). TP53 was found 9 times (23%), followed by 3 PI3KCA (8%), with several additional genes including (TERT, EGFR, MAP2K1, BRAF (non-V600E), IDH2, BRCA2, MYD88, CDH1, CTNNB1, SMO, NF1, and PTEN).
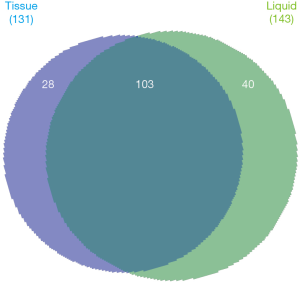
In order to determine if any of these alterations were acquired after anti-EGFR therapy, 12 patients were identified that had samples with MSAF >0 and had been treated with panitumumab or cetuximab in between tissue and liquid biopsy. In total, 50% (4 cetuximab and 2 panitumumab) of these patient samples were found to have alterations exclusively found in the liquid biopsy result and, thus, likely represented acquired resistance mutations. A CTNNB1 mutation was found in one case, and in another a CDH1 mutation was identified. Both alterations have been previously identified in patients with resistance to anti-EGFR therapy (14,15). Another patient sample had acquired KRAS G12V and EGFR S492R mutations, both of which have been shown to confer resistance (16,17). The remaining 3 patient samples had TP53 mutations, which have been shown to be associated with cetuximab resistance (18). These data demonstrate that in some cases, liquid biopsy can identify acquired resistance alterations that are not found in the primary tumor.
Fidelity between FoundationOne® and FoundationACT™ in matched liquid and tissue for the detection of rearrangements and copy number alterations
Only 4 rearrangements were identified in the dataset, and these were analyzed independently from the indel and substitution analysis. The rearrangements identified included 1 alteration found in both, 1 unique to tissue, and 2 unique to liquid biopsy. The two alterations found only in liquid included a BRCA1 rearrangement that did not have an identified partner and an EML1-ABL1 rearrangement. BRCA1 rearrangements have been identified in acquired resistance to platinum therapy (19) and this patient had been treated with platinum based-therapy. The ABL1-EML1 rearrangement has been associated with T-cell acute lymphoblastic leukemia (20). A single patient sample was found to have a CDH1-TANGO6 rearrangement, which was only detected by the tumor assay. This rearrangement has been described (21), but has not been associated as a prognostic or therapeutic marker. The rearrangement that was detected by both tissue and liquid was a TP53-TP53 intergenic rearrangement.
As liquid biopsy has shown high sensitivity for the detection of amplifications when the tumor fraction (MSAF) is >20% (5), we analyzed the dataset for amplifications in patient samples with MSAF >20%. There were 6 patient samples that each had a single amplification event. There was no concordance in detection of amplification events between tissue and liquid: 2 were found only in tissue (both MYC amplifications); to improve MYC amplifications detection in liquid biopsy, 2 additional baits covering SNPs in MYC have been added to the new liquid biopsy from Foundation Medicine (FoundationOne Liquid®). 4 amplifications were only found in the liquid biopsy (2 FLT3, 1 EGFR, and 1 MYC amplification); FLT3 and EGFR amplifications have been previously found in ctDNA as resistance mechanism (22).
Discussion
With the emergence of liquid biopsy as a diagnostic tool for clinicians to non-invasively analyze the molecular signature of patients’ tumors, it is critical to define how well liquid biopsy assays recapitulate the “gold-standard” of tissue-based diagnostics. The study described here is one of the largest studies of which we are aware in CRC that directly utilizes tissue and liquid biopsy from the same patients for comprehensive genomic analysis. Here we report the high fidelity between liquid and tissue biopsy in the genomic landscape of the colorectal cohort of patients. PPA was dependent on time between the tissue and liquid biopsy, where PPA of matched tissue and liquid biopsy taken within 30 days of each other was 100% and at 90 days was 95% for all short variants in patients with detectable ctDNA. PPA remained high (79%) even when tissue and liquid biopsy were taken 2 years apart, despite multiple lines of intervening therapy (Figure 1). We also analyzed KRAS, NRAS, and BRAF (NCCN recommend genes) independently as these genes have been recommended to be tested for in CRC and have therapeutic implications. Overall, PPA was similar for NCCN (80%) versus all (79%) alterations for the entire dataset but when analyzed in the <270 day cohort, PPA for NCCN genes increased to 90% (Table 7). We also analyzed the concordance of tissue and liquid biopsy for KRAS G12X for PPA, NPA, PPV, NPV, and OPA (Tables 5,6). Similarly to the other analysis, the <270 day cohort showed higher concordance: OPA (97%), PPA (93%), NPA (100%), PPV (100%), NPV (96%) (Table 6). The specimen in this cohort in with the single KRAS alteration that was not detected had a MSAF of 0.1%, suggesting very little circulating ctDNA.
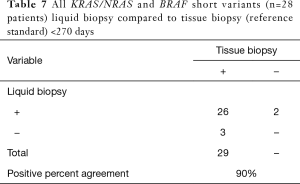
Full table
One of the critical concerns with liquid biopsy is the determination of whether the blood sample contains ctDNA. With the use of MSAF to determine the presence and relative amount of ctDNA in the samples, we found that approximately 20% of the liquid biopsy cases had no detectable ctDNA (MSAF =0). This is a similar level of detection of ctDNA in several other CRC cohorts (12,23-25). To better understand the dynamics of these cases with no evidence of ctDNA, we analyzed the tissue-based results, disease burden, and underlying therapy these patients received to determine if these factors may have contributed to these results. As therapy can have a large effect on the detection and quantity of ctDNA, we examined the clinical data for this variable. We found that 89% (16/18) of patients with MSAF =0 were on or had received therapy before the liquid biopsy; this was similar to the entire cohort (90%). In terms of disease stage, a higher proportion of samples with no ctDNA was present among the patients with lower stage disease [3 stage II (n=7, 42.9% with no ctDNA), 4 stage III (n=18, 22.2% with no ctDNA), and 11 stage IV (n=71, 15.5% with no ctDNA)]. We also saw that the MSAF was lower in early stage disease as compared to later stage disease (P<0.05) and a similar observation has also been reported by Bettegowda (12). Last, there was significant heterogeneity in sample collection that could have complicated the interpretation of the results. We did examine biopsies from metastatic versus primary tumor independently and neither was enriched for concordance with the liquid biopsy. We also stratified the patients by chemotherapy, targeted therapy, and radiation therapy before either the tumor or liquid biopsy and did not find any statistically significant differences in concordance with liquid biopsy; in some of these subsets the numbers were small making it unlikely to identify statistical differences. Future studies with a larger cohort with these subgroups are planned to address the limitations of this study.
MSAF is also a valuable tool for determining the performance of the assay, as it can serve as a surrogate for ctDNA fraction. As the analytical sensitivity of detecting alterations increases at higher tumor fractions, MSAF can be used to stratify cases for which the probability of detecting alterations is high or low. Liquid biopsy has high analytical sensitivity at MSAF 0.25–0.5% (95.8% sensitivity and 99.8% specificity for base substitutions and 87.7% sensitivity and 98.8% specificity for insertion and deletions), while tumor fractions below this have reduced sensitivity for detecting alterations (5). In 93% of the samples with ctDNA present (MSAF >0), MSAF was greater than 0.25%. When examining only cases with MSAF >0.25%, PPA between liquid and tissue results increased by 7% (86%) for indels/substitutions as compared to all cases with MSAF >0 (79%).
One of the major pre-analytical factors that affected concordance was time between the tumor biopsy and liquid biopsy. With less delay between tissue biopsy and liquid biopsy, the overall PPA was high for base substitutions and indels (100% PPA within 30 days, 95% PPA within 90 days, 87% PPA within 270 days). PPA decreased significantly when the liquid biopsy was taken more than 270 days after the tissue sample (P<0.05, 18% difference, 3.83–31.7%, 95% CI). In a subset analysis of each time group, the 0–30 day group (100% PPA) and the 30–90 day group (88% PPA) had high PPA; the later time points had variable PPA ranging from 44–86%. This consistent result across genes and across the cohort is not surprising, as greater delay between tissue biopsy and liquid biopsy means that, on average, patients would have received longer and more lines of therapy, radiation, and surgery, all of which could select for tumor cell populations or clones with genomic landscapes that are significantly divergent from the cells assayed in the tissue biopsy. These data have implications for future concordance studies of tissue and liquid biopsy and demonstrates the need for analysis to be restricted to samples collected contemporaneously (<30 days apart) without intervening therapy in order for meaningful comparison analysis to be performed.
Overall, in this study we show that genomic testing of liquid biopsy demonstrates high fidelity to tissue-based genomics in CRC. PPA is dependent on samples being collected temporally close—when samples are collected less than <90 days apart, PPA is >90% and remains >79% overall for short variants irrespective of time (up to 720 days) and therapy. Specific alterations (KRAS/NRAS and BRAF) showed similarity to tissue with a PPA of 90% in the <270 day cohort. The concordance of G12X was extremely high with overall percent agreement of 97%. Therefore, as liquid biopsy accurately represents the tumor genomic landscape of CRC, it can be a viable diagnostic tool for obtaining genomic information especially in cases where tissue is unavailable or biopsy in not feasible. This will become increasingly compelling as liquid biopsy incorporates microsatellite instability analysis to guide immunotherapy use.
In cases where tumor tissue profiling is not possible, these results provide compelling evidence that genomic profiling of ctDNA in late stage CRC shows a high concordance with tumor tissue sequencing results and can be used to identify most clinically relevant alterations capable of guiding therapy for these patients and has fidelity to the genomics of the tumor. However, although liquid biopsy offers a non-invasive modality for the molecular profiling of cancers and the clinical adoption of this method continues to increase, additional studies are still required to demonstrate clinical utility. This study demonstrates that future studies evaluating concordance should focus on concurrently-collected tissue and liquid biopsy samples whenever possible to evaluate fidelity. In general, patients currently receiving therapy, those with early stage disease, and/or those with low tumor burden may have lower or undetectable levels of ctDNA; thus, reliable ctDNA testing in the setting of tissue concordance studies, and in clinical practice, may be problematic.
Acknowledgments
None.
Footnote
Conflicts of Interest: G Li, D Pavlick, JH Chung, J He, M Cooke, J Hughes, SM Ali, M Nahas, VA Miller, P Stephens, B Forcier, JS Ross, AB Schrock are employees of Foundation Medicine, Inc. JP Gregg is a consultant/advisor and external speaker for FMI and on Speaker Bureau for AstraZeneca and BMS. The other authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by Western Institutional Review Board (Protocol No. 20152817).
References
- Benson AB 3rd, Venook AP, Cederquist L, et al. Colon Cancer, Version 1.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2017;15:370-98. [Crossref] [PubMed]
- Rankin A, Klempner SJ, Erlich R, et al. Broad Detection of Alterations Predicted to Confer Lack of Benefit From EGFR Antibodies or Sensitivity to Targeted Therapy in Advanced Colorectal Cancer. Oncologist 2016;21:1306-14. [Crossref] [PubMed]
- Sartore-Bianchi A, Trusolino L, Martino C, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol 2016;17:738-46. [Crossref] [PubMed]
- Zhao B, Wang L, Qiu H, et al. Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. Oncotarget 2017;8:3980-4000. [PubMed]
- Clark TA, Chung JH, Kennedy M, et al. Analytical Validation of a Hybrid Capture-Based Next-Generation Sequencing Clinical Assay for Genomic Profiling of Cell-Free Circulating Tumor DNA. J Mol Diagn 2018;20:686-702. [Crossref] [PubMed]
- Aaboud M, Aad G, Abbott B, et al. Search for the Decay of the Higgs Boson to Charm Quarks with the ATLAS Experiment. Phys Rev Lett 2018;120:211802. [Crossref] [PubMed]
- Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023-31. [Crossref] [PubMed]
- Richardson JT. The analysis of 2 x 2 contingency tables--yet again. Stat Med 2011;30:890-author reply 891-2. [Crossref] [PubMed]
- Chung JH, Pavlick D, Hartmaier R, et al. Hybrid capture-based genomic profiling of circulating tumor DNA from patients with estrogen receptor-positive metastatic breast cancer. Ann Oncol 2017;28:2866-73. [Crossref] [PubMed]
- Chen YH, Hancock BA, Solzak JP, et al. Next-generation sequencing of circulating tumor DNA to predict recurrence in triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy. NPJ Breast Cancer 2017;3:24. [Crossref] [PubMed]
- O'Leary B, Hrebien S, Morden JP, et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat Commun 2018;9:896. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004;304:554. [Crossref] [PubMed]
- Subbiah V, Khawaja MR, Hong DS, et al. First-in-human trial of multikinase VEGF inhibitor regorafenib and anti-EGFR antibody cetuximab in advanced cancer patients. JCI Insight 2017. [Crossref] [PubMed]
- Heindl S, Eggenstein E, Keller S, et al. Relevance of MET activation and genetic alterations of KRAS and E-cadherin for cetuximab sensitivity of gastric cancer cell lines. J Cancer Res Clin Oncol 2012;138:843-58. [Crossref] [PubMed]
- Hsu HC, Thiam TK, Lu YJ, et al. Mutations of KRAS/NRAS/BRAF predict cetuximab resistance in metastatic colorectal cancer patients. Oncotarget 2016;7:22257-70. [Crossref] [PubMed]
- Arena S, Bellosillo B, Siravegna G, et al. Emergence of Multiple EGFR Extracellular Mutations during Cetuximab Treatment in Colorectal Cancer. Clin Cancer Res 2015;21:2157-66. [Crossref] [PubMed]
- Hientz K, Mohr A, Bhakta-Guha D, et al. The role of p53 in cancer drug resistance and targeted chemotherapy. Oncotarget 2017;8:8921-46. [Crossref] [PubMed]
- Yamamoto KN, Hirota K, Takeda S, et al. Evolution of pre-existing versus acquired resistance to platinum drugs and PARP inhibitors in BRCA-associated cancers. PLoS One 2014;9:e105724. [Crossref] [PubMed]
- De Keersmaecker K, Graux C, Odero MD, et al. Fusion of EML1 to ABL1 in T-cell acute lymphoblastic leukemia with cryptic t(9;14)(q34;q32). Blood 2005;105:4849-52. [Crossref] [PubMed]
- Zhang J, White NM, Schmidt HK, et al. INTEGRATE: gene fusion discovery using whole genome and transcriptome data. Genome Res 2016;26:108-18. [Crossref] [PubMed]
- Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 2015;21:827. [Crossref] [PubMed]
- Morelli MP, Overman MJ, Dasari A, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol 2015;26:731-6. [Crossref] [PubMed]
- Strickler JH, Loree JM, Ahronian LG, et al. Genomic Landscape of Cell-Free DNA in Patients with Colorectal Cancer. Cancer Discov 2018;8:164-73. [Crossref] [PubMed]
- Schrock AB, Pavlick DC, Klempner SJ, et al. Hybrid capture-based genomic profiling of circulating tumor DNA from patients with advanced cancers of the gastrointestinal tract or anus. Clin Cancer Res 2018;24:1881-90. [Crossref] [PubMed]


