
Feasibility of circulating tumor DNA testing in hepatocellular carcinoma
Introduction
Exosomes, tumor educated platelets, circulating tumor cells, microRNA, and cell-free DNA (cfDNA) are all included in a group of circulating tumor related material, much of which is genomic in nature (1,2). Circulating tumor DNA (ctDNA) is, compared with cfDNA, tumor specific DNA in the circulation released after metabolic secretion, apoptosis or necrosis. Techniques have now been developed, with a number of assays commercially available to process plasma to extract ctDNA for any number of potential applications.
Detection and characterization of ctDNA is now possible and enables a multitude of clinical applications which are not reasonable options using routine tissue based sequencing methods (3). Potential applications include: (I) earlier detection of disease; (II) monitoring heterogeneity of tumors; (III) identification of targets for therapeutics; (IV) real-time therapeutic response assessment and earlier detection of tumor relapse; and (V) dynamic assessment of drug resistance development (3). Enabled by significant advancements in sequencing technology in recent years, investigations aimed at optimization of ctDNA technologies for use in clinical practice are ongoing.
Hepatobiliary malignancies including hepatocellular carcinoma (HCC) are poised to benefit greatly from ctDNA assay technologies given that (I) patients are often diagnosed with more advanced disease, (II) despite potentially curable interventions such as surgery, recurrences are common, (III) biopsies are not always available or often yield insufficient quantities of tumor cells and tumoral DNA for tissue-based genomic profiling. However, limited literature is available elucidating the feasibility of ctDNA testing in a real-time clinical setting in patients with HCC. Herein, we describe a cohort of such patients characterized using a clinically available ctDNA assay.
Methods
Patients
During the period extending from January 2015 to February 2018, 35 patients with HCC underwent ctDNA testing by Guardant Health (Redwood City, California, USA). All patients were seen at Mayo Clinic Cancer Center. Of the 35 patients, 45 samples were tested, with 6 patients having serial testing. Demographic information and date of blood collection were available for all patients. The analysis of data from patients in this cohort was approved by the Mayo Clinic institutional review board.
Comprehensive genomic testing in plasma
cfDNA was extracted from whole blood collected in Streck tubes, then shipped to a CLIA-certified, CAP-accredited laboratory (Guardant Health, Redwood City, California, USA). After double ultracentrifugation, 5–30 ng of cfDNA was isolated for digital sequencing. Leukocyte and tumor-derived cfDNA fragments, were simultaneously sequenced. The variant allele fraction (VAF) was calculated as the proportion of cfDNA harboring the variant in a background of wild-type cfDNA. The analytical sensitivity allowed detection of 1–2 mutant fragments in a 10-mL blood sample (0.1% limit of detection) with analytic specificity >99.9999%. Plasma copy number of 2.5–4.0 is reported as ++ amplification and >4.0 as +++, representing the 50th–90th and >90th percentile, respectively of all copy number alteration calls in the Guardant360 database.
Over the course of the study, the gene panel composition expanded from 54 to 68 to 70 to 73 genes. The currently utilized 73-gene panel includes the addition of 5 genes to and removal of 2 genes from the prior list. The vast majority (93%) of samples in this study were tested under the 73-gene panel.
Statistical analysis
The distribution of each continuous variable was reflected as its mean, standard deviation, and range. Categorical variables were depicted in terms of frequencies and percentages. Continuous variables were compared between groups by Wilcoxon rank sum test, and categorical data by Fisher exact test. All computations were carried out in SAS version 9.3 and R version 3.13.
Results
Data from a total of 35 patients were included in this study. A total of 44 samples were collected on these patients, with 3 patients having 2 tests performed and 3 patients having 3 tests performed. Patients had a median age of 66, with males representing 83% of the study population, and females making up 17%. Stage four disease was present in 43% of patients, with 23% of patients having stage three disease, and the rest having stages one and two disease. Sites of metastases varied, with most having involvement of either the adrenals or the lungs. Nodal metastases were uncommon, as was vascular invasion (Table 1).

Full table
Among all samples the median number of alterations, excluding variants of undetermined significance (VUS), per sample was 3.5 (range, 1–13), with a median allele frequency of 0.65% (range, 0.07–31.5%).
Testing was performed in patients prior to any therapy 34% of the time, in the middle of therapy treatment 9% of the time, and at the time of disease progression on therapy 57% of the time. Patients had received 0–3 prior systemic therapies overall, with 14 patients having received at least 1 prior systemic therapy. Patients had received prior locoregional therapy (LRT) (ablation, radioembolization, bland embolization or chemo embolization) 57% of the time, with ctDNA testing occurring a median of 269 days after (Table 1).
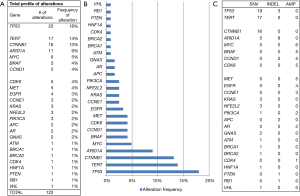
A total of 122 genetic alterations, excluding VUS or synonymous alterations, were seen. The profile of genetic alterations observed was broad (Figure 1A,B). The top 10 genes altered in this cohort of patients, excluding VUS or synonymous alterations, were TP53 (18%), TERT (14%), CTNNB1 (13%), ARID1A (9%), MYC (5%), BRAF (4%), CCND1 (4%), CDK6 (4%), and MET (4%), and EGFR (3%), thus just 5 genes being altered in ≥5% of patients, with many more less common alterations. Regarding the type of alterations, the majority (58%) were single nucleotide variations (point mutations), a minority (11%) were indels, and 31% were amplifications. No fusions included in the assay panel were detected (Figure 1C).
Discussion
Recently, analysis of TCGA data for 363 HCC cases by whole-exome sequencing and DNA copy number analyses, and 196 HCC cases by additional DNA methylation, RNA, miRNA, and proteomic expression analysis was reported (4). This analysis identified three subtypes of HCC, one of which was associated with a significantly poorer prognosis. Potential therapeutic targets were also indeed identified, for which agents currently exist, including alterations in WNT signaling, MDM4, MET, VEGFA, IDH1, and TERT. This data emphasizes the emerging importance of genomic profiling for patients with HCC. The standard for such profiling remains tissue-based testing. Unfortunately, tissue is very often not collected in patients with HCC given the ability to make the diagnosis based on imaging features alone. As such, ctDNA may represent a viable option for genomic profiling in this populating of patients.
Our data represent, to our knowledge, data from real-time, clinically collected ctDNA testing in the largest cohort of patients with HCC thus far reported in the literature. We describe the landscape of alterations, overall consistent with that seen in large tissue-based testing, such as the TCGA, including genes such as TP53, TERT and CTNNB1. Unfortunately, tissue was not available for genomic result comparison in a significant number of patients given that diagnosis of HCC is typically made based upon radiographic features, as invasive tissue biopsy poses a substantial bleeding risk. Furthermore, many of these plasma samples were drawn after intervention or at disease progression; therefore, tissue samples collected at diagnosis would not have reflected the cancer’s molecular makeup at the time of ctDNA analysis. One study showed that ctDNA and tissue next-generation sequencing (NGS) frequently showed different profiles, possibly related to the duration of time between tissue and blood samples [median =370 days (range, 29–876 days)] (5). Overall, the existing literature regarding ctDNA use in HCC is still quite limited, but our report of data herein suggests that application of ctDNA for genomic profiling of HCC is feasible and represents a tissue-free alternate in these often unbiopsied patients.
One of the most promising areas of potential for ctDNA regardless of cancer type has been its competency as a noninvasive, broad tool enabling precision medicine for some cancer patients. Potential exists in both its ability to identify therapeutic targets, and also to monitor disease response on treatment and detect emerging resistance. To enable its routine clinical use, proof of high concordance with tissue-based profiling, the current gold standard, will be necessary. There is a paucity of literature with regard to concordance of tissue and circulating DNA mutation analysis, unfortunately, especially in the realm of HCC. Complicating matters, this concordance may possibly vary depending upon the specific genomic alteration. One study reported data from 105 patients with gastrointestinal (GI) malignancies, including a cohort with HCC (number not reported). They revealed ctDNA-tissue concordance rates of 96%, 94%, 95%, and 91%, respectively, for four genomic alterations: KRAS amplification, MYC amplification, KRAS G12V, and EGFR amplification) (6). The overall concordance rate at the gene level for 68 genes included in both the tissue and liquid biopsy panels was 96%. Additionally, concordance across the 16 genes evaluable for amplifications in both ctDNA and tissue was 97%. Another study investigated whole exome sequencing and targeted deep sequencing of 32 multi-regional tumoral tissue samples from five HCC patients, compared with matched cfDNA. The genome profiling efficacy of cfDNA increased with sequencing depth, however their data suggested that tissue-based testing outperformed ctDNA (7). Another very small study of 3 HCC patients, focused on 574 cancer-related genes with recognized actionable mutations, evaluated the range of alterations and the corresponding ctDNA features to assess clinical significance. Analysis demonstrated ctDNA’s ability to overcome tumor heterogeneity and possibly to inform clinicians about tumor burden and prognosis (8). A fourth patient with samples from multiple tumor sites and sequential plasma samples identified 160 subclonal SNVs in tumor tissues. Plasma ctDNA impressively detected 97% of these tissue mutations (8).
Very limited data exists regarding the evaluation of the landscape of genomic alterations in HCC, as assessed by ctDNA. One study of 213 patients with advanced GI cancers described the applicability of ctDNA detection using a panel of 68 genes with a commercially available assay. HCC patients represented a small cohort of the study’s population (15%). Overall, 58% of patients had at least 1 non-VUS alteration, with a median number of alterations of 1. The number of detected alterations per patient differed by cancer type: 74% of patients with HCC had at least 1 characterized alteration. Among those 123 patients with characterized alterations, virtually all (>99%) had one or more alterations classified as potentially actionable. These observations suggest that many patients with GI cancers, including HCC, frequently have detectable and therapeutically actionable ctDNA alterations, detectable in a feasible manner (6). Another study reported ctDNA profiling data in 14 patients with HCC. All patients had somatic alterations with a median mutant allele fraction, 0.29% (range, 0.1–37.77%). Mutations were identified in several genes, similar to our study: “TP53 (57%), CTNNB1 (29%), PTEN (7%), CDKN2A (7%), ARID1A (7%), and MET (7%); amplification in CDK6 (14%), EGFR (14%), MYC (14%), BRAF (7%), RAF1 (7%), FGFR1 (7%), CCNE1 (7%), PIK3CA (7%), and ERBB2/HER2 (7%).” Eleven patients (79%) had more than 1 theoretically actionable alteration (9).
In sum, this pilot study demonstrates feasibility of ctDNA evaluation for patients with HCC and reports promising data. Further studies involving validation of ctDNA results with tissue-based profiling results, in addition to important biomarkers such as serum alpha fetoprotein levels in HCC, are needed to truly evaluate the generalizability of these initial preliminary observations.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The analysis of data from patients in this cohort was approved by the Mayo Clinic institutional review board.
References
- Ansari J, Yun JW, Kompelli AR, et al. The liquid biopsy in lung cancer. Genes Cancer 2016;7:355-67. [PubMed]
- Thierry AR, El Messaoudi S, Gahan PB, et al. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev 2016;35:347-76. [Crossref] [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Electronic address: wheeler@bcm.edu; Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327-41.e23. [Crossref]
- Ikeda S, Lim JS, Kurzrock R. Analysis of Tissue and Circulating Tumor DNA by Next-Generation Sequencing of Hepatocellular Carcinoma: Implications for Targeted Therapeutics. Mol Cancer Ther 2018;17:1114-22. [Crossref] [PubMed]
- Riviere P, Fanta PT, Ikeda S, et al. The Mutational Landscape of Gastrointestinal Malignancies as Reflected by Circulating Tumor DNA. Mol Cancer Ther 2018;17:297-305. [Crossref] [PubMed]
- Huang A, Zhao X, Yang XR, et al. Circumventing intratumoral heterogeneity to identify potential therapeutic targets in hepatocellular carcinoma. J Hepatol 2017;67:293-301. [Crossref] [PubMed]
- Cai ZX, Chen G, Zeng YY, et al. Circulating tumor DNA profiling reveals clonal evolution and real-time disease progression in advanced hepatocellular carcinoma. Int J Cancer 2017;141:977-85. [Crossref] [PubMed]
- Ikeda S, Tsigelny IF, Skjevik Å, et al. Next-Generation Sequencing of Circulating Tumor DNA Reveals Frequent Alterations in Advanced Hepatocellular Carcinoma. Oncologist 2018;23:586-93. [Crossref] [PubMed]


