
Fibroblast growth factor 7 signalling is disrupted in colorectal cancer and is a potential marker of field cancerisation
Introduction
Early diagnosis of colorectal cancer is crucial for achieving cure and is associated with improved survival (1). Molecular biomarkers that can identify mucosa at risk of neoplastic transformation, before the development of histological abnormality, hold the greatest promise. Based on the field cancerisation concept (2), the macroscopically normal mucosa around a cancer contains biological alterations, which occur early on in the cancer process (3). These changes contribute to neoplastic transformation without affecting the histological appearance of the mucosa. Characterisation of this ‘field defect’ around a cancer could enable identification of biomarkers of neoplastic risk (4).
Previous studies have investigated the role of tumour suppressor and oncogenes in field cancerisation; however, the importance of stromal cells in driving malignant epithelial cell growth has only recently come to the forefront of cancer research (5,6). Growth signals such as the fibroblast growth factors have particularly, been shown to contribute to epithelial cell proliferation in several cancers (7). Cellular proliferation and differentiation is governed by a family of some 22 fibroblast growth factors (8). Fibroblast growth factor 7 (FGF7) is a mitogen of mesenchymal origin that acts on a specific FGF receptor, FGFR2IIIb. Upon binding, it acts via the MAPK pathway to induce epithelial cell proliferation, migration and differentiation of epithelial cells (9). Studies in vitro have demonstrated that FGF7 contributes to wound repair and mucosal healing following a toxic injury to intestinal epithelial cells (10). To date there have been conflicting reports regarding its role in CRC formation with some authors proposing it is overexpressed in tumour tissue (11) whilst others indicate expression is no different compared with paired normal mucosa (12). Recent interest has focused on the role of its receptor, FGFR2b, which has been found to be overexpressed in colorectal cancer, suggesting a putative role in governing growth of malignant cells (13). In contrast, there have been some reports where FGFR2b expression has been linked with a less aggressive tumour type (14). Thus, it is unclear how FGF7-FGFR2 signalling contributes to CRC formation.
To address this further, this study utilised a novel approach by taking serial samples along the colon enabling the expression level at the tumour site to be compared to that found at distant sites. A separate cohort of control subjects was included to determine the expression level when there was no mucosal abnormality in the colon. Thus, the purpose of this study was twofold; firstly, to ascertain the importance of FGF7 and its receptor FGFR2 as an early molecular marker in field defects around CRC; secondly, downstream FGF7 targets in the tumour and mucosal field around a cancer were evaluated to determine how FGF7-FGFR2 signalling contributes to CRC formation.
Methods
Participants
All participants were provided with written information about the study. Written informed consent was gained. The study was performed in accordance to the Declaration of Helsinki. Ethical approval was acquired from Coventry and Warwick Local Research Ethics Committee (MREC ref No. 09/H1211/38) as well as Research Governance approval and sponsorship from the University Hospital Coventry & Warwickshire Research & Development office.
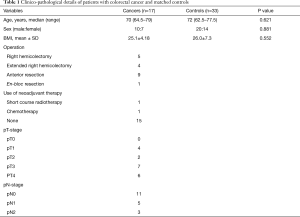
In total, 51 patients (21 females) were recruited, of which, 17 had cancer. There were two patients with synchronous lesions therefore 19 cancers were analysed. The clinicopathological details for the control subjects and cancer patients are given in Table 1; there were no significant differences between the two groups. Each cancer patient was age and sex matched to two control subjects. Control subjects were patients undergoing colonoscopy with no endoscopic or histological evidence of mucosal abnormality. Colorecl cancer patients were matched to control subjects depending upon the site of tumour/polyp; left sided tumours to rectal samples and right sided tumours to caecal samples.
Full table
Mucosal pinch biopsies were taken from MNM of the caecum and rectum at time of endoscopy in control subjects. In CRC patients, mucosal biopsies were taken immediately after bowel division from the colectomy specimen prior to fixation in formalin. Mucosal tissue was taken from the resection margin, tumour site and adjacent to the tumour (within 1 cm). Once retrieved, all tissue samples were placed immediately in RNA later (Life Technologies, UK). Liquid nitrogen was used to snap freeze the tissue samples which were then stored at −80 °C.
Extraction of RNA and purity
The mucosal biopsy tissue (~0.2 mg) was utilised to extract RNA with a column-based isolation method (RNeasy Mini Tissue Kit: Qiagen, UK) according to the manufacturer’s instructions. This yielded 30 µL RNA, from which genomic DNA was removed using a DNase I Kit (Sigma). Three point five µL (1,000 U/mL) DNase I digestion enzyme combined with 3.5 µL reaction buffer were added at room temperature for 15 min, after which 3.5 µL stop solution (50 mM EDTA) was added. This was then centrifuged (up to 8 s), heated (to 70 °C for 10 min) and subsequently chilled on ice. A spectrophotometer (Nanodrop, Labtech, UK) was used to quantify the RNA by measuring the absorbance at 260 nm using duplicate samples (1.5 µL). In order to determine RNA purity, the ratio was calculated between absorbance at 260/280 nm and at 260/230 nm. Only RNA samples with values between 1.8 and 2.1 were utilised for experiments.
Synthesis of complimentary (cDNA)
A Bioline kit (No. BIO-65026) was used to synthesise complimentary DNA (cDNA). The following were added to a 200 µL sterile microcentrifuge tube: 1 µL 10 mM dNTP mix (Invitrogen, UK), 1 µL random hexamers 50–250 ng (Bioline, UK), 250 ng RNA and RNAse free water to make up a 10 µL solution.
Samples were spun and heated to 70 °C (for 10 min) and chilled on ice (further 2 min). Ten µL reverse transcription mastermix was added to the samples (1 µL RNase inhibitor, 4 µL of 5× reaction buffer, 0.5 µL reverse transcriptase (200 U/µL) with RNase free water to make up a volume of 10 µL). This resulted in in a final total volume of 20 µL. Following incubation at room temperature (5 min), the samples were placed in a thermocycler (Biorad, UK). Each sample was heated for 5 min at 37 °C, for 55 min at 42 °C and for 15 min at 70 °C to generate the required cDNA.
Quantitative real-time polymerase chain reaction (qRT-PCR)
To undertake qRT-PCR, an ABI 7500 standard Sequence Detection System (Applied Biosystems, UK) was used. In a 96 well plate, reactions were primed in 25 µL volumes containing 1 µL cDNA, 12.5 µL Taqman universal PCR mastermix (Applied Biosystems, UK) and TaqmanTM Gene Expression Assay (Applied Biosystems, UK).
In each reaction, an endogeneous control was used which was the housekeeping gene 18 S (ribosomal RNA). Triplicate samples were processed in order to improve accuracy of results. Each sample was heated to 50 °C (2 min), 95 °C (10 min) and 95 °C for 15 seconds over 40–44 cycles, followed by 60 °C (1 min). To measure gene expression, commercially available Taqman primers were utilised [Applied Biosystems, UK, FGFR2 (Hs01552918_m1) and FGF7 (Hs00940253_m1)]. For all reactions, the primers for the housekeeping and target gene were placed together in the same well. ΔCt was calculated by deducting Ct of the housekeeping gene from Ct of the target gene. ΔCt for each sample was calculated by averaging across the triplicates. The inverse of the ΔCt is a measure of the level of gene expression and was utilised in statistical analysis. In order to determine differences in gene expression between groups, the relative fold change was calculated using the formula 2−ΔΔCt where ΔΔCt is the difference between the ΔCt for the control sample and ΔCt for the test sample, which could either be the cancer/polyp sample, the resection margin or the tissue adjacent to the cancer/polyp (15).
Extraction of protein
Protein was extracted from the colonic samples that had been snap frozen. After homogenising each tissue sample, 1 mL of protein lysis buffer was added. This contained 5 mL of 1× radioimmunoprecipitation (Millipore, UK) in which 100 µL of protease and phosphatase inhibitors [8 mg sodium fluoride (Fisher Scientific, UK) and 20 mg sodium vanadate (Acros Organics) in 2 mL 1× RIPA with 2 Roche Complete Mini protease inhibitor cocktail tablets)] were dissolved. The Bio-Rad detergent compatible protein assay kit (Bio-Rad Laboratories, CA, USA) was used to measure protein concentration. A series of known dilutions of bovine serum albumin (BSA, fraction V, Sigma, UK) (2 µg/µL) were utilised to construct a standard curve of protein concentration against absorbance. Finally, a spectrophotometer (Tecan, UK) was used to measure absorbance at a 595 mm wavelength to enable calculation of the protein concentration (µg/µL).
Western blot analysis
This was performed using a 7.5–10% polyacrylamide gel (Geneflow Ltd, Fradley, UK) for which 25–30 µg of protein was loaded onto the gel. As per protocol, the proteins were resolved over 60–90 min using SDS-PAGE by applying a current at 100 V. The proteins were transferred to Immobilon-P transfer membranes 0.45 µm pore size (Fisher Scientific, UK). Initially, the membrane was incubated in 0.2% I-Block PBS-tween (PBST) for 60 min. It was then transferred into an incubator at 4 °C overnight containing primary rabbit-derived antibody (Cell Signalling & Abcam, UK). Measurement of β-actin expression ensured equal amounts of protein were loaded. After washing in PBST, membranes were incubated in either horseradish peroxidase antibody produced in goat or anti-rabbit IgG (whole molecule), IgG fraction of antiserum and buffered aqueous solution (Sigma No. A9169, USA). Visualisation of protein bands was achieved using chemiluminescence with ECL/ECL+ (GE Healthcare, UK) and exposure on hyperfilm MP (Fisher Scientific, UK). Protein expression was estimated by measuring intensity of exposure with densitometry (GeneTool software, Syngene, UK). All values for protein expression were normalised by subtracting the intensity of exposure obtained for β-actin.
Statistical calculations
All statistical calculation and analysis were performed using SPSS version 22 (SPSS Inc, Chicago, Illinois, USA). Raw mRNA expression values were log10 transformed to normalise the data prior to statistical analysis. The unpaired t-test was used to compared the expression levels between cancer patients and control subjects. Comparison of gene or protein expression amongst samples taken at different points along the colon in the same colorectal cancer patient was achieved using the paired t-test. A P value of <0.05 was considered significant.
Results
Dysregulation of FGF7 gene expression
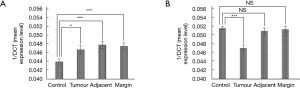
In patients with colorectal cancer, there was upregulation of FGF7 at multiple points along the colon (see Figure 1) including the resection margin (∆∆CT 21.1), tumour tissue itself (∆∆CT 21.4) and its adjacent MNM (∆∆CT 20.9). This was in contrast to control subjects (∆∆CT 22.6). However, its receptor, FGFR2 was significantly down regulated only in the tumour tissue (∆∆CT 21.2 in tumour tissue compared to ∆∆CT 19.4 for controls, P<0.001). Conversely, expression of FGFR2 at the resection margin (∆∆CT 19.4) and in the adjacent MNM (∆∆CT 19.6) did not differ when compared to control subjects. Using the paired t test, comparison was made of samples taken along the colon in individual colorectal cancer patients. This showed significant downregulation of both FGF7 (P=0.036) and FGFR2 (P=0.013) in the tumour tissue compared to the resection margin.
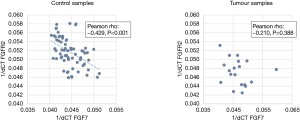
There was negative correlation between FGF7 and FGFR2 gene expression in control subjects (see Figure 2) but no correlation was not observed in tumour tissue.
Akt pathway key for FGF7-FGFR2 signalling axis
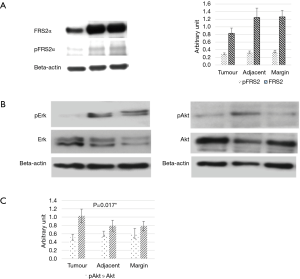
Western blot analysis demonstrated reduced protein expression of FRS2α and phospho-FRS2α in tumour tissue compared to samples taken from the resection margin. This would suggest that the downstream effects of FGF7 signalling was reduced only at the tumour site. In addition, there was also a reduction in Akt protein expression but no differences were noted in Erk 1/2 expression (Figure 3A,B,C). Protein expression levels were similar in tissues taken from the resection margin and tissue adjacent to tumour.
Discussion
This study has identified FGF7 as a putative biomarker of CRC field cancerisation utilising a well-matched cohort of cancer patients and control subjects. The use of serial samples taken along the colectomy specimen enabled changes in signalling of the FGF7-FGFR2 axis to be elucidated. FGFR2 and its downstream targets, FRS2α and Akt were downregulated in the tumour samples indicating that malignant transformation is accompanied by loss of FGF7 activity through the Akt and not the Erk pathway. This highlights that at the tumour site, cellular differentiation is lost (Akt pathway) whilst proliferation (Erk pathway) is preserved.
There have been several reports highlighting that FGF7 acts as a mitogen and therefore would be expected to augment epithelial cell growth. However, studies investigating tissue expression in colorectal cancer have been inconclusive in elucidating the precise role of FGF7 in tumour formation (11,12,14). Our study has demonstrated distinct differences in gene expression of FGF7-FGFR2 that occur only in tumour tissue and not at distant colonic sites. The correlation between FGF7 and FGFR2 gene expression noted in control subjects was lost at the tumour site. An alteration in FGFR trafficking may be responsible for this observation and may accompany malignant transformation. Upon activation, in a physiological environment, FGFRs undergo endocytosis which decreases the number of receptors present in the membrane. Thus, through negative feedback, FGFR activity is autoregulated; disruption of this process can result in disordered growth factor acitivty contributing to cancer formation (16). Similarly, others have shown that well differentiated colorectal tumours and those with shallow wall invasion have greater FGFR2 expression (14) compared to poorly differentiated tumours implying that in aggressive tumours, the FGFR2 signal is lost. Examples of other cancers where FGFR2 expression is lost include cancers of the salivary gland, bladder and prostate (17-19). This implies that the FGF7-FGFR2 axis may actually protect against toxic injury and neoplastic transformation.
Our study investigated this hypothesis further by measuring the protein expression of certain FGF7 downstream targets. We found that Akt protein expression is reduced at the tumour site only whilst Erk expression is maintained. Despite being activated by FGF7, these two pathways have been shown to play distinct roles. A study on pancreatic duct cells demonstrated that the MEK-Erk 1/2 pathway was important for proliferation and the Akt pathway played a role in cell differentiation (20). Hence, in our study with colorectal cancer, results suggest that FGF7 may have lost its ability to regulate cell differentiation as Akt expression was reduced. However, activity via the MEK-Erk 1/2 pathway was preserved suggesting that cell proliferation was retained. Others have shown that the effects of FGF7 on cell growth in vitro may differ depending upon whether it is a cancer cell or normal epithelial cell. FGF7 could not stimulate tumour cells to proliferate but was a strong mitogen for normal epithelial cells (12). Similarly, only primary, immortalised keratinocytes could by stimulated to grow by recombinant FGF7; it had no effect upon malignant head and neck squamous cancer cells (21). If FGF7 activity is blocked by a neutralising antibody, cell proliferation of normal keratinocytes is blocked; there is no effect on malignant cells. Unlike normal cells, malignant cells did not express FGFR2b. Therefore, FGF7 could not stimulate cell proliferation in a paracrine manner as it was able to in normal cells. Taken together, our results imply that it may not be the ability of FGF7 to drive cell proliferation that is important but rather, the loss of its ability to regulate cell differentiation that could contribute to malignancy.
Due to the limitations in tissue availability, our study was unable to evaluate expression of the specific isoform of the FGFR2 receptor. There is emerging evidence proposing that the transformation of isoforms for example from isoform FGFR2IIIb to isoform FGFR2IIIc could be associated with a more aggressive cancerous phenotype. Such a class switch may be important in driving the malignant process and holds potential for therapeutic agents in the future (22). At present, the precise role of this receptor transition in colorectal cancer is unclear and given the changes in gene expression we observed, it was felt that it was more important to investigate downstream signalling rather than receptor class switch.
In conclusion, we have demonstrated that FGF7 is upregulated in patients with colorectal cancer compared with control subjects. From these findings, we postulate that FGF7 plays a role in field cancerisation and could be utilised in a screening population to identify those patients with colorectal cancer or in those with cancer or polyps, to ultimately help elucidate who is at most risk of further neoplasia. A further novel finding is that signalling along the FGF7/FGFR2 cascade is disrupted only in the tumour tissue and not in tissue taken adjacent to the tumour. Thus, it is possible that reduced FGF7 activity contributes directly to the neoplastic process or that the malignant cells develop mutations that inactivate this pathway. Regardless of whether the changes observed are a cause or consequence of malignancy, specific directed therapy (chemo or nutraceutical) which has the ability to restore the balance of FGF7 would be welcome.
Acknowledgements
We thank late Mr Joseph Bench of Coventry for his invaluable donation to the Colorectal Cancer Research Fund at University Hospitals Coventry and Warwickshire NHS Trust, which enabled this research to be conducted. We are grateful to the patients treated at University Hospital Coventry & Warwickshire NHS Trust for their altruism.
Funding: The Bowel Disease Research Foundation and the Colorectal Cancer Research Fund at University Hospitals of Coventry and Warwickshire NHS Trust.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: All participants were provided with written information about the study. Written, informed consent was gained. The study was performed in accordance to the Declaration of Helsinki. Ethical approval was acquired from Coventry and Warwick Local Research Ethics Committee (MREC ref No. 09/H1211/38) as well as Research Governance approval and sponsorship from the University Hospital Coventry & Warwickshire Research & Development office.
References
- NCIN. National Cancer Intelligence Network. Retrieved September 2015. Available online: http://www.ncin.org.uk/cancer_information_tools/eatlas/
- Patel A, Tripathi G, Gopalakrishnan K, et al. Field cancerisation in colorectal cancer: a new frontier or pastures past? World J Gastroenterol 2015;21:3763-72. [Crossref] [PubMed]
- Dakubo GD, Jakupciak JP, Birch-Machin MA, et al. Clinical implications and utility of field cancerization. Cancer Cell Int 2007;7:2. [Crossref] [PubMed]
- Rubin H. Fields and field cancerization: the preneoplastic origins of cancer: asymptomatic hyperplastic fields are precursors of neoplasia, and their progression to tumors can be tracked by saturation density in culture. Bioessays 2011;33:224-31. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Dotto GP. Multifocal epithelial tumors and field cancerization: stroma as a primary determinant. J Clin Invest 2014;124:1446-53. [Crossref] [PubMed]
- Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 2010;10:116-29. [Crossref] [PubMed]
- Ornitz DM, Itoh N. The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip Rev Dev Biol 2015;4:215-66. [Crossref] [PubMed]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev 2005;16:139-49. [Crossref] [PubMed]
- Cai YJ, Wang WS, Liang HY, et al. Keratinocyte growth factor up-regulates Interleukin-7 expression following intestinal ischemia/reperfusion in vitro and in vivo. Int J Clin Exp Pathol 2012;5:569-80. [PubMed]
- Watanabe M, Ishiwata T, Nishigai K, et al. Overexpression of keratinocyte growth factor in cancer cells and enterochromaffin cells in human colorectal cancer. Pathol Int 2000;50:363-72. [Crossref] [PubMed]
- Otte JM, Schmitz F, Banasiewicz T, et al. Expression of keratinocyte growth factor and its receptor in colorectal cancer. Eur J Clin Invest 2000;30:222-9. [Crossref] [PubMed]
- Matsuda Y, Ishiwata T, Yamahatsu K, et al. Overexpressed fibroblast growth factor receptor 2 in the invasive front of colorectal cancer: a potential therapeutic target in colorectal cancer. Cancer Lett 2011;309:209-19. [Crossref] [PubMed]
- Yoshino M, Ishiwata T, Watanabe M, et al. Keratinocyte growth factor receptor expression in normal colorectal epithelial cells and differentiated type of colorectal cancer. Oncol Rep 2005;13:247-52. [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Auciello G, Cunningham DL, Tatar T, et al. Regulation of fibroblast growth factor receptor signalling and trafficking by Src and Eps8. J Cell Sci 2013;126:613-24. [Crossref] [PubMed]
- Diez de Medina SG, Chopin D, El Marjou A, et al. Decreased expression of keratinocyte growth factor receptor in a subset of human transitional cell bladder carcinomas. Oncogene 1997;14:323-30. [Crossref] [PubMed]
- Naimi B, Latil A, Fournier G, et al. Down-regulation of (IIIb) and (IIIc) isoforms of fibroblast growth factor receptor 2 (FGFR2) is associated with malignant progression in human prostate. Prostate 2002;52:245-52. [Crossref] [PubMed]
- Amann T, Bataille F, Spruss T, et al. Reduced expression of fibroblast growth factor receptor 2IIIb in hepatocellular carcinoma induces a more aggressive growth. Am J Pathol 2010;176:1433-42. [Crossref] [PubMed]
- Uzan B, Figeac F, Portha B, et al. Mechanisms of KGF mediated signaling in pancreatic duct cell proliferation and differentiation. PLoS One 2009;4:e4734. [Crossref] [PubMed]
- Hille A, Grüger S, Christiansen H, et al. Effect of tumour-cell-derived or recombinant keratinocyte growth factor (KGF) on proliferation and radioresponse of human epithelial tumour cells (HNSCC) and normal keratinocytes in vitro. Radiat Environ Biophys 2010;49:261-70. [Crossref] [PubMed]
- Matsuda Y, Hagio M, Seya T, et al. Fibroblast growth factor receptor 2 IIIc as a therapeutic target for colorectal cancer cells. Mol Cancer Ther 2012;11:2010-20. [Crossref] [PubMed]


