
Natural killer NKG2A and NKG2D in patients with colorectal cancer
Introduction
Colorectal cancer (CRC), the third most commonly diagnosed cancer in the world, has a higher predominance in developed countries (1,2). According to the Egypt National Cancer registry, the incidence rates/100,000 population of individual cancer sites are: in Upper Egypt in 2008 were 6.2 and 9.6, respectively; in Middle Egypt incidences were 6.7 and 9.7, respectively; while in Lower Egypt values were 8.0 and 10.7, respectively for both males and females (3).
Although, there have been well established tumor factors that affect the prognosis in CRC patient (T stage, N stage, residual tumor following surgery with curative intent, and tumor grade) (4). However, there are multiple determinants for development and survival of tumor. One of these, are host responses which are realized to have a major responsibility (5).
Natural killer (NK) cells, large granular lymphocytes (LGLs), play a principal role in primary defense against viruses and tumors. NK cells have both activating and inhibitory receptors. They adjust their activity through an equilibrium between the inhibitory and stimulatory signal cascades. Natural-killer group 2 member A (NKG2A) as an inhibitory receptor and D (NKG2D) as an activating receptor, belong to the C-type lectin superfamily in NK cells (6-8).
The NKG2D ligands (NKG2DLs) consist of a various assemblage of proteins that are concomitant in structure to MHC class I. Several studies indicate that NKG2D and NKG2DLs engagement promotes NK cells expansion, survival, and cytotoxic activity (9,10). Thus, NKG2D/NKG2DLs interaction constitutes an important start to activate the strong immune response against malignant cells (11).
NKG2D ligands are not demonstrated on normal cells, however on solid tumors and few types of leukemia through induction of cancer-related pathways and oncogenes (12,13). Hence, owing to the loss of these ligands in normal cells and their expression in different types of cell stress as viral infections and cancer, NKG2D is considered as one of molecules involved in immune surveillance (14,15).
Moreover, NKG2A/NKG2D and their ligands interaction have been associated to a diverse physiologic and pathologic functions. Meanwhile, efforts have been done trying to explain their roles in the regulation of immune cells activities and responses (15). Hence, the objective of the current study was to detect NKD2A and NKG2D mRNA expression in serum NK cells acquired from peripheral blood of metastatic, non-metastatic CRC patients, and healthy individuals.
Methods
A case control study conducted on 36 patients with CRC, as well as from 15 healthy individuals (group matched). Patients with CRC of all stages were enrolled from the Gastroenterology Oncology Department, Ain Shams University Hospitals. The current study was carried out over a 7 months period (February 2017 to August 2017). The CRC patients were further divided into 23 non-metastatic CRC (group 1) and 13 metastatic CRC (group 2). The mean age of patients was 45.9±16.0. All patients have pathological proved diagnosis of colorectal adenocarcinoma, ≥18 years old and didn’t receive chemotherapy and/or radiotherapy for their cancer. None of the patients had colorectal tumors other than adenocarcinoma or synchronous malignancy, major organ failure, primary or acquired immunodeficiency, presence of any systemic infection (clinical manifestations and proved by diagnostic investigations) or on any immunosuppressant therapy. The study was approved by the Oncology review board and Faculty of Medicine Ain Shams University research ethical and Human rights committee. All participants assigned an informed consent before taking part.
Study design
The participants were assessed by detailed medical history and clinical examination. Complete blood picture with differential count, renal and hepatic functions were performed for all participants. Full colonoscopy and biopsy, histological examination of colonoscopic biopsy and tumor marker [carcinoembryonic antigen (CEA)] were done for all patients. Radiological assessment of patients was carried out by computerized tomography of the abdomen and pelvis with contrast, and/or magnetic resonance imaging, positron emission tomography scan for proper initial staging.
RNA extraction and cDNA synthesis
Total RNA was extracted from peripheral blood mononuclear cells (PBMCs) using the RNeasy Mini Kit (QIA amp mini kit, Qiagen, USA), that combines the selective binding properties of a silica-based membrane with the speed and convenience of micro-spin technology, a specialized high-salt buffering system allows RNA to bind to the QIAamp membrane. cDNA was synthesized by incubating 10 µL total RNA (contain up to 1 µg RNA) in a 20 µL reaction mixture consist of RT Buffer, and RT Primer Mix. The entire reaction takes place at 42 °C and is then inactivated at 95 °C using QuantiTect® Reverse Transcription kit (QIAGEN, USA).
Real time polymerase chain reaction (RT-PCR)
The primers used for NKG2A, NKG2D, and β-actin are shown in Table 1. All the primers were synthesized and validated by (Qiagen, Germany). Reactions were performed in a total volume of 20 µL, which included 5 µL of cDNA sample, 5 µL of 0.8 µM primer pair, and 10 µL of 2X PCR mix (Qiagen, Germany). PCR was performed as follows: 10 min at 95 °C and 45 cycles of 30 s at 95 °C, 45 s at 60 °C, and 30 s at 72 °C. Incubation and on-line detection of the PCR products were carried out using optical 96-well plates and the LightCycler 480 sequence detection system (Qiagen, Germany). All runs contained positive, negative, and endogenous controls. RT-PCR positive samples were identified automatically, when the fluorescence signal exceeded the threshold level determined by the machine. RQ value was estimated. RQ =2−ΔΔCt where ΔΔ Ct = [Ct (tested gene) − Ct (endogenous control)] – [mean Ct (control) − Ct (endogenous control)] which was performed by the computer software as the mean of the control group was used to calculate the fold change for miRNA in the study group according to Livak and Schmittgen method (16).
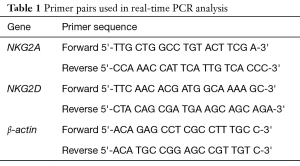
Full table
Statistical methodology
Analysis of data was performed using IBM© SPSS© Statistics version 23 (IBM© Corp., Armonk, NY, USA) and MedCalc© version 15.8 (MedCalc© Software bvba, Ostend, Belgium). The Shapiro-Wilk test was used to detect numerical data distribution. Unpaired t-test was used to compare normally distributed numerical variables (mean ± SD) and inter-group differences. Either Mann-Whitney test (two-group comparison) or the Jonckheere-Terpstra trend test (multiple-group comparison) was used to compare non-normally distributed numerical data (median and interquartile) and intergroup differences. Post hoc comparison was detected by Conover test after the Jonckheere-Terpstra trend test if needed. The Bonferroni method was use to correct the P value for multiple post hoc comparisons with the Conover test. Categorical variables were presented as ratio or number and percentage and differences were compared using Fisher’s exact test. Interpretation of correlation coefficient (Spearman rho) which is used for correlations was as follows: <0.2, 0.2–0.39, 0.4–0.59, 0.6–0.79, 0.8–1.0 as very weak, weak, moderate, strong, very strong respectively.
Diagnostic/predictive value of NKG2D or NKG2A was assessed by receiver-operating characteristic (ROC) curve analysis. The area under the ROC curve (AUC) interpretation was as follows: 0.9–1.0 excellent, 0.8–0.89 good, 0.7–0.79 fair, 0.6–0.69 poor, <0.6 fail. For post-hoc comparisons with the Conover test, a P value <0.01 (statistically significant) (Bonferroni correction). Otherwise, P values <0.05 (statistically significant).
Results
Demographic and laboratory characteristics of all participants
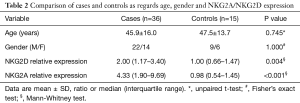
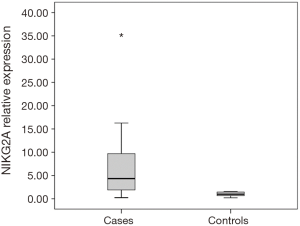
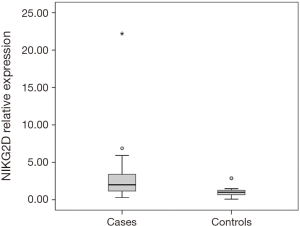
Thirty-six patients with CRC and 15 healthy individuals were enrolled in the study. As regards, either age or gender, no statistical significant difference between studied groups was detected. Serum NKG2D and NKG2A relative expression were significantly higher in patients compared to the control group with P=0.004, and <0.001 respectively (Table 2). NKG2A expression in patients and controls was shown in Figure 1 and NKG2D expression in patients and controls was shown in Figure 2. The median (interquartile range) for CEA was 3.0 (1.4–12.2), for NKG2D relative expression was 2.00 (1.17–3.40) and for NKG2A relative expression was 4.33 (1.90–9.69).
Full table
Diagnostic accuracy of NKG2D or NKG2A for differentiation between patients and healthy individuals
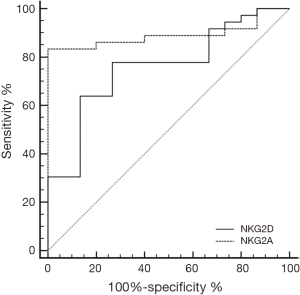
ROC curve analysis for discrimination between patients and controls using NKG2D or NKG2A showed sensitivity (77.8, 83.33%) and specificity (73.33, 100%), with calculated area under the curve (AUC) of 0.759, 0.891 respectively and 95% CI, (0.832 to 0.979) (Figure 3).
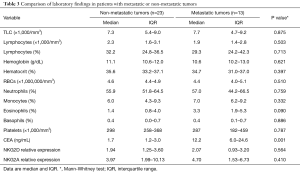
Comparison of laboratory findings in metastatic or non-metastatic CRC
No statistical significant difference between patients with metastatic and non-metastatic CRC was detected regarding complete blood picture, renal function, liver function and NKG2D or NKG2A expression however statistical significant increase in CEA in metastatic compared to non-metastatic with P value =0.001 as shown in Table 3.
Full table
Discussion
NK cells occupy a pivotal role in innate immunity against tumors. The integration of signals from inhibitory and activating receptors regulate NK cells functions (17). Little is known about the role of NKG2A/NKG2D as vital inhibitory and activating receptors, respectively, in CRC (18). The NKG2D receptor are normally expressed in human on NK cells, γδ T cells, CD8 T cells, some autoreactive and immunosuppressive CD4 T cells (19,20).
Engagement of NKG2D with its ligands evokes protective cytolytic response with release of cytokines. These can be obtained either by activation directly or via NK cells and CD8 T cells co stimulation (21,22). NKG2D ligands are extensively demonstrated on a variety of cancer cells and primary solid tumors (23). Additionally, human NKG2D ligands, are expressed when cells experience physical stress or any changes due to genetic variation (24).
The molecular mechanism underlying NKG2D regulation is not fully established. NKG2D expression can be induced by several cytokines including IL-2, IL-7 and IL-15. IL-15 and TNF are the most significant biologically activating cytokines, however NKG2D expression can be inhibited by IL-21, TGF-β and macrophage migration inhibitory factor (MIF) (25). The rationale behind the current study was to determine NKG2D as well as NKG2A expression in patients with CRC.
Our study was conducted on 36 patients with CRC, as well as 15 matched healthy individuals. The patients were further classified into two groups: 23 non-metastatic CRC (group 1) and 13 metastatic CRC (group 2).
Our study revealed that expression of NKG2D and NKG2A mRNA were significantly higher in patients compared to the control group with P=0.004, <0.001 respectively. This was surprisingly in contrast with the several previous studies that investigated NKG2 receptors in CRC.
In a study carried by Shen and coworkers, they assessed NKG2A and NKG2D expression by real-time PCR. They found no statistical significant difference regarding NKG2A expression levels in both control group and CRC patients, however NKG2D expression levels were significantly lower in the CRC patients compared to healthy individuals. Hence, their results detected an imbalance in NKG2A/NKG2D expression in CRC patients (18). In addition, Doubrovina and colleagues have previously examined the expression of NKG2D in colon cancer patients and detected their decrease in comparison with healthy individuals (26).
In the current study, no statistical significant difference between patients with metastatic and non-metastatic CRC was detected regarding complete blood picture, renal function, liver function and NKG2D or NKG2A expression however statistical significant increase in CEA in metastatic compared to non-metastatic.
In contrast, Gharagozloo et al. observed that the percentages of NKG2D+NK cells and their mRNA expression were decreased during tumor progression, indicating a significant decrease in metastatic colon cancer patients (27).
Actually, different studies display various changes in percentage of serum NK cells in diverse cancers. In one study on colon cancer, all patients without metastasis showed a significant expansion of NK cells in the peripheral blood when compared to healthy controls, while patients with metastatic CRC did not show significant difference in NK-cells number when compared to non-metastatic patients (26). It is accepted that tumors have the ability to develop different ways to escape immunity through an immunoediting process (28).
Cancers could evade NKG2D mediated immune responses by various mechanisms. Tumors can down-regulate NKG2D expression either by shedding of soluble NKG2D ligands or secretion of immunosuppressive cytokines as TGF-beta (29-31). In addition, continuous expression of NKG2D ligands can provoke downregulation of NKG2D expression (14,32,33). However, the actual mechanisms that regulate the immunity against cancer seemed to be more complex and in need to many studies to further explore the role of each element. Major histocompatibility complex class I-related chain A (MICA) (the most important ligand for NKG2D receptor) was an independent good prognostic marker in CRC for stage I and II, but not later stage cancers (23) as well as in pancreatic cancer (34).
Besides, MICA/B and UL-16 binding proteins (ULBP2) were associated with decreased relapse in early-stage breast cancer (35). MICA/B (miR-106b) micro RNA was decreased in primary ovarian cancer and increased in CRC, gastric cancer, and hepatocellular carcinoma (HCC) (36-38). Hence, different studies of microRNA expression in human cancers had shown contradictory results.
A tumor-specific expression pattern of MICA has been noticed in a wide spectrum of epithelial tumor cells, as melanoma, colon, breast, lung, ovary and renal. However, Zhang and colleagues examined the expression of MICA and found MICA association with HCC especially advanced stage, and MICA expression was decreased in HCC tissues when compared with noncancerous tissues (39).
Our study showed an elevation of circulating inhibitory NKG2A more than circulating excitatory NKG2D, though they were both elevated in cancer patients more than controls. This clearly draws our attention that the balance between both receptors does not follow that simple scheme of up and down regulation and the regulation of such relationship passes through more complex mechanisms that are needed to be explored.
NKG2D ligands may induce another tumor survival mechanism other than promoting tumor immune evasion and suppression. In a conceptual twist, different parts of ex vivo breast, ovarian, prostate, and colon cancer cells express signaling expert NKG2D, thus augmenting the presence of its ligands for self-stimulation of tumor growth and possibly malignant development (40).
Cancer cell NKG2D, when targeted by engagement of ligands on adjacent cancer cells, activates the oncogenic PI3K-AKT-mammalian target of rapamycin (mTOR) signaling axis. This is commonly found to be hyperactive in cancer, and downstream effectors controlling cell growth and protein synthesis or antibody crosslinking. Besides, as in lymphocytes, targeting of NKG2D stimulates c-Jun N-terminal kinase (JNK) and extracellular signal-regulated protein kinase (ERK) phosphorylation in mitogen-activated protein kinases (MAPK) cascades (40). These kinases are activation targets of receptor tyrosine kinases as the epidermal growth factor receptor (EGFR), which frequently cause excessive cancer cell expansion with increased motility and survival because of mutation of aberrant expression (41).
Hence, NKG2D itself might represent the main factor underlying poor clinical outcomes that have been associated with cancer cell expression of its ligands (34,38,42,43). In spite of their protective role in tumor immune surveillance, NKG2D and its ligands could be misapplied as tumor survival device, allowing immune evasion and suppression, and quite possibly stimulation of tumor growth and progression. The uncertain role of NKG2D as an oncoprotein still needs various in vivo experimental studies (25).
Intensive research should be held to investigate the exact role of NKG2D in tumorigenesis and whether it has a distinct and well-established role in tumor immunity or it is just a gear in a highly complicated machinery that regulate and control tumor growth. Among points of strength in our study is that it is one of the first research work that could detect an increase of NKG2D in CRC comparison to healthy controls, and this finding opens wide field of research to investigate the exact mechanism of regulation and control of NKG2 receptors in malignancy. Limitations of the study include small sample size, and that we did not evaluate the whole mechanism of the NKG2 receptors regulation by measuring the receptors ligands as well.
Conclusions
Our work is one of the first research work that could detect an increase in NKG2D and NKG2A in CRC in comparison to healthy controls. Indeed, more research should investigate the regulatory mechanisms of NK cells and their receptors that are involved in tumorigenesis. This would to better understand the details of immunity machinery against tumor cells, and other ways the later could escape body immune surveillance.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the Oncology review board and Faculty of Medicine Ain Shams University research ethical and Human rights committee (No. FMASU R: 77/2018). All participants assigned an informed consent before taking part.
References
- Des Guetz G, Uzzan B, Morere JF, et al. Duration of adjuvant chemotherapy for patients with non-metastatic colorectal cancer. Cochrane Database Syst Rev 2010;20:CD007046. [PubMed]
- Adelstein BA, Macaskill P, Chan SF, et al. Most bowel cancer symptoms do not indicate colorectal cancer and polyps: a systematic review. BMC Gastroenterol 2011;11:65. [Crossref] [PubMed]
- Ibrahim AS, Khaled HM, Mikhail NN, et al. Cancer incidence in egypt: results of the national population-based cancer registry program. J Cancer Epidemiol 2014;2014:437971. [Crossref] [PubMed]
- Compton CC, Lawrence Cooper HS, Stanley R, et al. American joint committee on cancer prognostic factors consensus conference, Arch Patho lab Med 2000;124:979-94.
- Roxburgh CS, McMillan DC. Cancer and systemic inflammation: treat the tumour and treat the host. Br J Cancer 2014;110:1409-12. [Crossref] [PubMed]
- Coudert JD, Held W. The role of the NKG2D receptor for tumor immunity. Semin Cancer Biol 2006;16:333-43. [Crossref] [PubMed]
- Eagle RA, Jafferji I, Barrow AD. Beyond stressed self: evidence for NKG2D ligand expression on healthy cells. Curr Immunol Rev 2009;5:22-34. [Crossref] [PubMed]
- Bottino C, Castriconi R, Moretta L, et al. Cellular ligands of activating NK receptors. Trends Immunol 2005;26:221-6. [Crossref] [PubMed]
- Pende D, Cantoni C, Rivera P, et al. Role of NKG2D in tumor cell lysis mediated by human NK cells: cooperation with natural cytotoxicity receptors and capability of recognizing tumors of nonepithelial origin. Eur J Immunol 2001;31:1076-86. [Crossref] [PubMed]
- Moretta A, Bottino C, Vitale M, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol 2001;19:197-223. [Crossref] [PubMed]
- Watson NF, Spendlove I, Madjd Z, et al. Expression of the stress–related MHC class I chain–related protein MICA is an indicator of good prognosis in colorectal cancer patients. Int J cancer 2006;118:1445-52. [Crossref] [PubMed]
- Gasser S, Orsulic S, Brown EJ, et al. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature 2005;436:1186-90. [Crossref] [PubMed]
- Boissel N, Rea D, Tieng V, et al. BCR/ABL oncogene directly controls MHC class I chain–related molecule A expression in chronic myelogenous leukemia. J Immunol 2006;176:5108-16. [Crossref] [PubMed]
- Coudert JD, Zimmer J, Tomasello E, et al. Altered NKG2D function in NK cells induced by chronic exposure to NKG2D ligand-expressing tumor cells. Blood 2005;106:1711-7. [Crossref] [PubMed]
- Guerra N, Tan YX, Jonker NT, et al. NKG2D deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity 2008;28:571-80. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Lanier LL. NK cell receptors. Annu Rev Immunol 1998;16:359-93. [Crossref] [PubMed]
- Shen Y, Lu C, Tian W, et al. Possible association of decreased NKG2D expression levels and suppression of the activity of natural killer cells in patients with colorectal cancer. Int J Oncol 2012;40:1285-90. [Crossref] [PubMed]
- Nausch N, Cerwenka A. NKG2D ligands in tumor immunity. Oncogene 2008;27:5944-58. [Crossref] [PubMed]
- Dai Z, Turtle CJ, Booth GC, et al. Normally occurring NKG2D+CD4+ T cells are immunosuppressive and inversely correlated with disease activity in juvenile-onset lupus. J Exp Med 2009;206:793-805. [Crossref] [PubMed]
- González S, Groh V, Spies T. Immunobiology of human NKG2D and its ligands. Curr. Top. Microbiol. Immunol 2006;298:121-38. [Crossref] [PubMed]
- Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol 2008;9:495-502. [Crossref] [PubMed]
- McGilvray RW, Eagle RA, Watson NF, et al. NKG2D ligand expression in human colorectal cancer reveals associations with prognosis and evidence for immunoediting. Clin Cancer Res 2009;15:6993-7002. [Crossref] [PubMed]
- Groh V, Rhinehart R, Randolph-Habecker J, et al. Costimulation of CD8 T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat Immunol 2001;2:255-60. [Crossref] [PubMed]
- El-Gazzar A, Groh V, Spies T. Immunobiology and Conflicting Roles of the Human NKG2D Lymphocyte Receptor and its Ligands in Cancer. J Immunol 2013;191:1509-15. [Crossref] [PubMed]
- Doubrovina ES, Doubrovin MM, Vider E, et al. Evasion from NK cell immunity by MHC class I chain-related molecules expressing colon adenocarcinoma. J Immunol 2003;171:6891-9. [Crossref] [PubMed]
- Gharagozloo M, Kalantari H, Rezaei A, et al. The decrease in NKG2D+ Natural Killer cells in peripheral blood of patients with metastatic colorectal cancer. Bratisl Lek Listy 2015;116:296-301. [PubMed]
- Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol 2004;22:329-60. [Crossref] [PubMed]
- Groh V, Wu J, Yee C, et al. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature 2002;419:734-8. [Crossref] [PubMed]
- Castriconi R, Cantoni C, Della Chiesa M, et al. Transforming growth factor beta 1 inhibits expression of NKp30 and NKG2D receptors: consequences for the NK-mediated killing of dendritic cells. Proc Natl Acad Sci U S A 2003;100:4120-5. [Crossref] [PubMed]
- Lee JC, Lee KM, Kim DW, et al. Elevated TGF-beta1 secretion and down-modulation of NKG2D underlies impaired NK cytotoxicity in cancer patients. J Immunol 2004;172:7335-40. [Crossref] [PubMed]
- Wiemann K, Mittrucker HW, Feger U, et al. Systemic NKG2D down-regulation impairs NK and CD8 T cell responses in vivo. J Immunol 2005;175:720-9. [Crossref] [PubMed]
- Oppenheim DE, Roberts SJ, Clarke SL, et al. Sustained localized expression of ligand for the activating NKG2D receptor impairs natural cytotoxicity in vivo and reduces tumor immunosurveillance. Nat Immunol 2005;6:928-37. [Crossref] [PubMed]
- Duan X, Deng L, Chen X, et al. Clinical significance of the immunostimulatory MHC class I chain-related molecule A and NKG2D receptor on NK cells in pancreatic cancer. Med Oncol 2011;28:466-74. [Crossref] [PubMed]
- de Kruijf EM, Sajet A, van Nes JG, et al. NKG2D ligand tumor expression and association with clinical outcome in early breast cancer patients: an observational study. BMC Cancer 2012;12:24-35. [Crossref] [PubMed]
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006;103:2257-61. [Crossref] [PubMed]
- Dahiya N, Sherman-Baust CA, Wang TL, et al. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS One 2008;3:e2436. [Crossref] [PubMed]
- Li Y, Tan W, Neo TW, et al. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci 2009;100:1234-42. [Crossref] [PubMed]
- Zhang J, Xu Z, Zhou X, et al. Loss of expression of MHC class I-related chain A (MICA) is a frequent event and predicts poor survival in patients with hepatocellular carcinoma. Int J Clin Exp Pathol 2014;7:3123-31. [PubMed]
- Benitez AC, Dai Z, Mann HH, et al. Expression, signaling proficiency, and stimulatory function of the NKG2D lymphocyte receptor in human cancer cells. Proc Natl Acad Sci USA 2011;108:4081-6. [Crossref] [PubMed]
- Normanno N, De Luca A, Bianco C, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006;366:2-16. [Crossref] [PubMed]
- Wu JD, Higgins LM, Steinle A, et al. Prevalent expression of the immunostimulatory MHC class I chain-related molecule is counteracted by shedding in prostate cancer. J Clin Invest 2004;114:560-8. [Crossref] [PubMed]
- Rebmann V, Schutt P, Brandhorst D, et al. Soluble MICA as an independent prognostic factor for the overall survival and progression-free survival of multiple myeloma patients. Clin Immunol 2007;123:114-20. [Crossref] [PubMed]


