|
Original Article
Exosome can prevent RNase from degrading microRNA in feces
Yoshikatsu Koga1, Masahiro Yasunaga1, Yoshihiro Moriya2, Takayuki Akasu2, Shin Fujita2, Seiichiro Yamamoto2, Yasuhiro
Matsumura1
1Investigative Treatment Division, Research Center for Innovative Oncology, National Cancer Center Hospital East, Kashiwa; 2Department of Surgery,
National Cancer Center Hospital, Tokyo, Japan
Corresponding to: Yasuhiro Matsumura, MD, PhD. Investigative Treatment
Division, Research Center for Innovative Oncology, National Cancer
Center Hospital East, 6-5-1 Kashiwanoha, Kashiwa 277-8577, Japan. Tel:
+81-4-7134-6857; Fax: +81-4-7134-6857. E-mail: yhmatsum@east.ncc.go.jp.
|
|
Abstract
Background: Because the stability of miRNA in feces has not been clarified, we examined the stability of miRNA in
feces.
Methods: RNase was added into culture media of HT-29 cells and fecal homogenates. The relative quantifications of
miRNA were analyzed by real-time RT-PCR.
Results: Cellular miRNA or exosomal miRNA were protected from RNase by the cellular membrane or the exosome;
meanwhile, free miRNA was degraded immediately and completely by RNase.
Conclusion: The present study revealed that exosome or cellular membrane could prevent RNase from degrading miRNA
inside the exosome or cells even in a dreadful condition, as in feces.
Key words
exosome, colonocyte, fecal miRNA test, miRNA, cancer screening
J Gastrointest Oncol 2011; 2: 215-222. DOI: 10.3978/j.issn.2078-6891.2011.015
|
|
Introduction
MicroRNAs (miRNAs), which are small (18-25
nucleotides) noncoding RNA molecules, regulate the
activity of specific mRNA targets and play a major role in
cancer. The function of miRNA is the downregulation of
multiple target gene expressions by degrading the mRNA
or blocking its translation into protein through RNA
interference ( 1, 2). The let-7, miR-34 family, miR-126,
miR-143, miR-145, and the miR-200 family are considered
to be tumor suppressor miRNAs in colorectal cancer (CRC)
( 3-7). Because the expression level of tumor suppressor
miRNAs in cancer tissue was lower than in normal tissue,
these tumor suppressor miRNAs may become candidates for future miRNA-based cancer therapy ( 8). On the other
hand, since the expression level of the oncogenic miRNAs,
such as miR-17-92 cluster, miR-21, and miR-135, in cancer
tissue was higher than in normal tissue, these oncogenic
miRNAs could be used for a marker of prognosis or poor
response to chemotherapy ( 9-14). Exosomes are nanoparticles, 50-100 nm in diameter,
and are released from cells into extracellular matrixes
through fusion of multivesicular bodies with the plasma
membrane ( 15, 16). Recent reports indicate that miRNAs
are circulating stably in bloodstream wrapping in exosomes,
which can prevent RNase from degrading the miRNAs
( 17-21). Therefore several methods for miRNA-based
early cancer detection using serum, plasma, and urine
are reported ( 21-23). Also, several studies are available of
the possible use of the miRNA-based method for CRC
screening in serum ( 24, 25) and in feces ( 26). We have been developing new screening methods for
CRC by applying molecular biological tools to exfoliated
colonocytes isolated from naturally evacuated feces ( 27-29).
In the past few years especially, we have reported the fecal
RNA test, including the CRC-related gene expression
analysis ( 30) and the CRC-related miRNA expression
analysis ( 31). Within this context, we investigate the stability of miRNA in feces.
|
|
Materials and Methods
Cell line and fecal samples
The human colorectal cancer cell line HT-29 (American
Tissue Culture Collection, Rockville, MD) was cultured
in the Dulbecco’s Modified Eagle Medium (DMEM),
supplemented with 10% fetal bovine serum (FBS), 100 U/
mL penicillin G, 100 μg/mL streptomycin, and 0.25 μg/mL
amphotericin B at 37°C in a humidified atmosphere of 5%
CO2: 95% air.
Naturally evacuated fecal samples were obtained from
3 healthy volunteers with no endoscopical abnormalities.
Volunteers were instructed to evacuate at home into a
disposable 5 × 10-cm polystyrene tray (AsOne, Osaka,
Japan) and to bring the fecal sample to our laboratory at
4oC. The samples were then immediately prepared for the
next step.
Isolation of exfoliated colonocytes from feces using EpCAM
beads
EpCAM (epithelial cell adhesion molecule) beads (JSR,
Tsukuba, Japan), immunomagnetic beads conjugated with
EpCAM monoclonal antibody (mAb) (clone B8-4), were
used for isolation of colonocyte from feces ( 32). Fecal samples were processed as described previously
( 28). Brief ly, one gram of fecal sample was homogenized
with a buffer (40 mL) consisting of Hanks’ solution, 10%
fetal bovine serum (FBS), and 25 mM HEPES buffer
(pH 7.35) at 200 rpm for 1 min using a Stomacher system
(Seward, Thetford, UK). The homogenate was filtered
through a nylon filter (pore size, 512 μm), and following
the addition of 40 μL of EpCAM beads, the sample mixture
was incubated for 30 min under gentle rolling conditions
at room temperature. The mixture on the magnet was
incubated on a shaking platform for 15 min at room
temperature. The supernatant was then removed, and the
colonocytes in the pellet were stored at −80°C until RNA
extraction.
Isolation of exosome from culture media or feces using CD63
beads
CD63 beads (JSR), immunomagnetic beads conjugated
with CD63 mAb (R&D systems, Minneapolis, MN),
were used for isolation of exosome from culture media or
feces.
Ten microliters of CD63 beads were applied to 1 mL
of culture media of HT-29 cells, and the sample mixture
was incubated for 30 min under gentle rolling conditions
at room temperature. The mixture on the magnet was incubated on a shaking platform for 15 min at room
temperature. The supernatant was then removed, and the
exosomes in the pellet were stored at −80°C until RNA
extraction.
Isolation of exosome from feces was processed in the
same manner as described above. The exosomes isolated
from feces using CD63 beads were stored at −80°C until
RNA extraction.
Extraction of total RNA
Fecal samples were homogenized as described previously
( 33, 34), and total RNA was extracted from all homogenates
using a miRNeasy Mini Kit (Qiagen, Valencia, CA), in
accordance with the manufacturer’s instructions. Brief ly,
one gram of feces was homogenized with 5 mL of Isogen
(Nippon Gene, Toyama, Japan), using an IKA Ultra-Turrax
homogenizer (IKA-Werke, Staufen, Germany) at 6,000 rpm
for 2 min. The homogenates were centrifuged at 15,000 rpm
for 5 min at 4°C. The supernatants were transferred into a
new tube, up to 5 mL more Isogen was added, and 1.5 mL of
chloroform was then added. HT-29 cel ls, exosomes isolated by CD63 beads,
and colonocytes isolated by EpCAM beads were also
homogenized with 1 mL of Isogen, and to the homogenates
0.2 mL of chloroform were added.
One milliliter of culture media was also homogenized
with 3 mL of Isogen-LS (Nippon gene), and to the
homogenates 0.2 mL of chloroform were added.
All of the tubes were shaken vigorously for 30 sec,
and centrifuged at 15,000 rpm for 15 min at 4°C. The
aqueous phase was transferred into a new tube. One-anda-
half volume of 100% ethanol was added, and the tube
was vortexed for 15 sec. The mixture was poured onto a
miRNeasy spin column (Qiagen), and the columns were
centrifuged at 10,000 rpm for 15 sec at room temperature.
The rema ining s teps were done according to the
manufacturer's instructions. Each sample was eluted in 100
μL of RNase-free water. The total RNA was electrophoresed
using a Cosmo-I microcapillary electrophoresis (Hitachi
High-Technologies, Tokyo, Japan), and the concentrations
of total RNA was determined using a NanoDrop UV
spectrometer (LMS, Tokyo, Japan). The RNA samples were
stored at −80°C until analysis.
cDNA synthesis and real-time RT-PCR
For miRNA expression analysis, cDNAs for U6, miR-16,
and miR-21 were synthesized. For this purpose, we used the
commercially available TaqMan MicroRNA Assay (Applied
Biosystems, Foster, CA).
cDNA for miRNA was synthesized using a TaqMan
MicroRNA RT Kit (Applied Biosystems) in accordance with the manufacturer’s instructions. The reaction mixture
consisted of 2 μL (or 5 ng) of total RNA, 0.5 μL of 10 ×
RT buffer, 1 μL of 5 × RT primer, 0.05 μL of dNTPs (100
mM), 0.06 μL of RNase Inhibitor (20 U/μL), and 0.33 μL
of MultiScribe Reverse Transcriptase (50 U/μL) in a final
reaction volume of 5 μL.
The reaction mixture for real-time PCR consisted of 4 μL
of a template cDNA, 10 μL of TaqMan Fast Universal PCR
Master Mix (Applied Biosystems), and 1 μL of 20 × primer/
probe mixture in a total reaction volume of 20 μL. Real-time
RT-PCR was performed with precycling heat activation at
95°C for 20 sec, followed by 40 cycles of denaturation at 95°
C for 3 sec and annealing/extension at 60°C for 30 sec in an
Applied Biosystems 7500 Fast Real-Time PCR System.
Susceptible to RNase degradation
To evaluate the susceptibility to RNase, RNA extracted
from HT-29 cells was treated using RNase (Qiagen,
final concentration: 5 μg/mL) at 4°C or 37°C for 0, 5,
10, 20, and 30 min. After the treatment, all samples
were electrophoresed using a Cosmo-I microcapillary
electrophoresis, the concentrations of total RNA were
evaluated using a NanoDrop UV spectrometer, and the
expressions of miRNA from HT-29 cells were analyzed
using real-time RT-PCR.
Analysis of RNA protection from RNase
HT-29 cells (5 × 105 cells) were plated into a 10-cm cell
culture plate (Corning, Corning, NY). After an exchange
for 10 mL of fresh medium the next day, HT-29 cells were
cultured for 48 hr. The HT-29 cells were then incubated at
37°C for 0, 30, 60, and 90 min after addition of RNase (final
concentration, 5 μg/mL). The culture media and cells were
processed as described above, and free miRNA, exosomal
miRNA, and cellular miRNA could be obtained. Three
replicates were performed in each sample.
One gram of fecal sample from 3 volunteers was put into
Stomacher Lab Blender Bags (Seward) and incubated at 37°
C for 0, 30, 60, and 90 min after the addition of RNase (final
concentration, 5 μg/mL). The fecal samples were processed,
and fecal miRNA, exosomal miRNA, and colonocyte
miRNA could be obtained as described above.
Statistical analysis
The miRNA expression analyses were conducted using
the comparative Ct (threshold cycle) method. The relative
quantification for each miRNA was analyzed using a twosided
t-test. Statistical analyses were performed using
StatView Ver. 5 for Windows (Abacus Concepts, Berkeley,
CA). P<0.05 was considered statistically significant.
|
|
Results
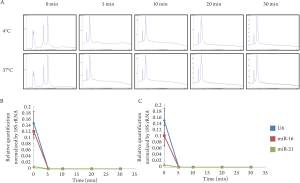
Degradation of naked RNA from HT-29 cells using RNase
Total RNA extracted from HT-29 cells was treated, using
5 μg/mL of RNase, and electrophoresed. Two peaks, 18S
and 28S ribosomal RNA (rRNA), were observed in the total
RNA without treatment of RNase ( Fig 1A). On the other
hand, no rRNA peak was observed in the total RNA treated
with RNase. Small RNAs, including miRNA or degrading
RNA, were observed in all samples. miRNA expressions
treated with RNase at 4°C were significantly lower than
those without treatment (U6: P=0.002; miR-16: P=0.0006;
miR-21: P=0.003) ( Fig 1B). Same as above, miRNA
expressions treated with RNase at 37°C were significantly
lower than those without treatment (U6: P=0.003; miR-16:
P=0.006; miR-21: P=0.01) ( Fig 1C). As a consequence,
naked RNA was degraded by 5 μg/mL of RNase at both 4°C
and 37°C for only 5 min.
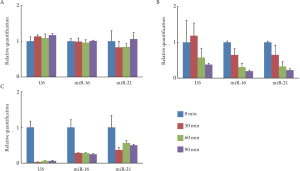
miRNA protected by exosome or cellular membrane from
RNase in HT-29 cells
To examine how miRNA was protected from RNase in
vitro, we cultured HT-29 cells in the medium containing
RNa se; cellular miRNA extracted from the cells,
exosomal miRNA from the exosomes, and free miRNA
from the culture media were then analyzed. Cellular
miRNA was sufficiently conserved under the treatment
of RNase for 90 min ( Fig 2A). Exosomal miRNA was
conserved under the treatment of RNase for 30 min;
however, the miRNA was degraded thereaf ter ( Fig
2B). Free miRNA was degraded by the treatment of
RNase within 30 min ( Fig 2C). Cellular miRNA was
sufficiently protected from RNase by cellular membrane.
Exosomal miRNA was partially protected by exosome.
On the other hand, free miRNA in the culture media was
degraded immediately by RNase.
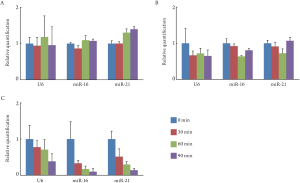
Effects of RNase on miRNA in exosome or colonocyte in feces
We also examined the susceptibility of miRNA to RNase
degradation in feces. Colonocyte miRNA extracted from
the fecal colonocyte, exosomal miRNA extracted from the
fecal exosomes, and fecal miRNA extracted from the fecal
homogenates were analyzed. Ct values of U6 in colonocyte
miRNA, exosomal miRNA, and fecal miRNA without
treatment of RNase were 31.14 (26.57-36.13) (mean
(range)), 33.23 (30.40-35.15), and 32.60 (31.08-34.29),
respectively ( Table 1). Ct values of miR-16 were 28.60
(25.71-30.83), 29.69 (28.79-31.01), and 30.36 (29.47-31.05),
respectively. Also, Ct va lues of miR-21 were 27.23
(23.83-29.00), 27.92 (26.27-30.46), and 29.32 (28.16-30.68),
respectively. Colonocyte miRNA and exosomal miRNA were not susceptible to RNase degradation ( Fig 3A and 3B).
On the other hand, fecal miRNA was degraded efficiently by
the treatment of RNase ( Fig 3C). In the feces, miRNA was
sufficiently protected from RNase by cellular membrane
and exosome.
|
|
Discussion
In the clinical samples, RNA is degraded rapidly by RNase
existing in any body fluids such as sweat, sputum, or blood.
The effects of RNase should be, therefore, considered in
the RNA-based analysis on clinical samples. Although
several storage buffers inhibiting the effect of DNase and
RNase were available, we have been investigating the
CRC screening method based on the analysis using the
colonocytes isolated from feces. In our preliminary study,
the colonocytes could not be isolated from feces stored
in the storage buffers. Therefore we have investigated the suitable storage condition of fecal samples for our screening
test.
Recently it was reported that miRNA was secreted from
tumor cells via exosome and was transported to endothelial
cells by paracrine induction ( 35). This indicates that
exosome is not only a secretory tool, but that it also supports
miRNA. We have been investigating the CRC screening
method ( 30, 31). And then we thought that fecal miRNA
(free miRNA) from fecal homogenates, exosomal miRNA
from fecal exosomes, and colonocyte miRNA from fecal
colonocytes might be candidates for the fecal miRNA test.
Exfoliated colonocytes were isolated from feces by EpCAM
beads, using a previously published method ( 28, 32).
Exosomes could be isolated using both the centrifugation
method ( 19, 35) and the cell isolation method by anti-
CD63 mAb conjugated immunomagnetic beads ( 36) In the
present study, HT-29 cells cultured in the media containing
RNase were analyzed, and fecal homogenates were treated by RNase. Although free miRNA (fecal miRNA) was
degraded rapidly, cellular miRNA (colonocyte miRNA)
was highly conserved. In the culture media, exosomal
miRNA was conserved for a 30-min treatment of RNase,
but degraded for a 90-min treatment. On the other
hand, the fecal exosome could be conserved for a 90-min
treatment of RNase. These indicated that cellular membrane
prevented RNase from degrading miRNA in cells, but that
the exosome partially prevented RNase from degrading
miRNA in exosome. In this study, U6, miR-16, and miR-21 were analyzed
because U6 and miR-16 were used for internal control as an expression of miRNA in several reports ( 31, 37)
and miR-21 was one of the miRNAs important for CRC
carcinogenesis ( 38, 39). The expression of miR-21 in the
CRC tissue was higher than that in the normal colorectal
mucosa; however, no significant difference was seen
between the early stage of CRC and the advanced stage of
CRC regarding the expression of miR-21 ( 31). Recently
fecal-based RNA tests have been noticed because of their
simplicity and cost-effectiveness ( 33, 34, 40), however, fecal
miRNA was unstable under the existence of RNase. For
the clinical use of fecal miRNA, it was therefore necessary
to store the fecal sample under strict conditions. On the other hand, exosomal miRNA or colonocyte miRNA were
protected from RNase by exosome or cellular membrane.
This information may be important for the clinical use of
fecal miRNA in future CRC mass screening. In the present
study, we examined miRNA protection from RNase in fecal
samples precisely and could show that exosomal miRNA is
more stable than free miRNA in a deadful condition like in
feces. |
|
Funding
This work was supported by a Grant-in-Aid for the Program
for Promotion of Fundamental Studies in Health Sciences of
the National Institute of Biomedical Innovation (NIBIO) of
Japan (to Y. Koga); Young Scientists (B) from the Ministry
of Education, Culture, Sports, Science, and Technology of
Japan (to Y. Koga); the Innovation Promotion Program from
the New Energy and Industrial Technology Development
Organization (NEDO) of Japan (to Y. Matsumura); and the Regional Innovation Cluster Program (City Area Type) (to
Y. Matsumura).
|
|
Acknowledgments
We thank Professor Shigeru Kanaoka (Hamamatsu
University School of Medicine) for his advice on fecal RNA
extraction and Dr. Satoshi Katayose (JSR Corp.) for his
technical support concerning immunomagnetic beads. We
are also grateful to Mr. Suguru Fujisawa, Mr. Yohei Hisada,
Ms. Satoe Miyaki, and Ms. Noriko F. Abe for their technical
assistance, and to Ms. Kaoru Shiina for her secretarial
assistance.
|
|
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and
function. Cell 2004;116:281-97.[LinkOut]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259-69.[LinkOut]
- Slaby O, Svoboda M, Michalek J, Vyzula R. MicroRNAs in colorectal
cancer: translation of molecular biology into clinical application. Mol
Cancer 2009;8:102.[LinkOut]
- Shi B, Sepp-Lorenzino L, Prisco M, Linsley P, deAngelis T, Baserga R.
Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the
growth of colon cancer cells. J Biol Chem 2007;282:32582-90.[LinkOut]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng
A, et al. RAS is regulated by the let-7 microRNA family. Cell
2005;120:635-47.[LinkOut]
- Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, et al. Role of
miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene
2009;28:1385-92.[LinkOut]
- Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, Guda K. The
noncoding RNA, miR-126, suppresses the growth of neoplastic cells by
targeting phosphatidylinositol 3-kinase signaling and is frequently lost
in colon cancers. Genes Chromosomes Cancer 2008;47:939-46.[LinkOut]
- Trang P, Medina PP, Wiggins JF, Ruffino L, Kelnar K, Omotola M, et al.
Regression of murine lung tumors by the let-7 microRNA.. Oncogene
2010;29:1580-7.[LinkOut]
- Yamamichi N, Shimomura R, Inada K, Sakurai K, Haraguchi T, Ozaki
Y, et al. Locked nucleic acid in situ hybridization analysis of miR-21
expression during colorectal cancer development. Clin Cancer Res
2009;15:4009-16.[LinkOut]
- Monzo M, Navarro A, Bandres E, Artells R, Moreno I, Gel B, et al.
Overlapping expression of microRNAs in human embryonic colon and
colorectal cancer. Cell Res 2008;18:823-33.[LinkOut]
- Diosdado B, van de Wiel MA, Terhaar Sive Droste JS, Mongera
S, Postma C, Meijerink WJ, et al. MiR-17-92 cluster is associated
with 13q gain and c-myc expression during colorectal adenoma to
adenocarcinoma progression. Br J Cancer 2009;101:707-14.[LinkOut]
- Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA,
Bolijn A, et al. Regulation of the adenomatous polyposis coli gene by the
miR-135 family in colorectal cancer. Cancer Res 2008;68:5795-802.[LinkOut]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et
al. MicroRNA expression profiles classify human cancers. Nature
2005;435:834-8.[LinkOut]
- Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs):
ammunition to target miRNAs implicated in human disease? Gene Ther
2006;13:496-502.[LinkOut]
- van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a
common pathway for a specialized function. J Biochem 2006;140:13-21.[LinkOut]
- Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from
biogenesis and secretion to biological function. Immunol Lett
2006;107:102-8.[LinkOut]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived
exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol
2008;110:13-21.[LinkOut]
- Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection
of microRNA expression in human peripheral blood microvesicles.
PLoS One 2008;3:e3694.[LinkOut]
- Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T.
Secretory mechanisms and intercellular transfer of microRNAs in living
cells. J Biol Chem 2010;285:17442-52.[LinkOut]
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA,
Hopmans ES, Lindenberg JL, et al. Functional delivery of viral miRNAs
via exosomes. Proc Natl Acad Sci U S A 2010;107:6328-33.[LinkOut]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-
Agadjanyan EL, et al. Circulating microRNAs as stable bloodbased
markers for cancer detection. Proc Natl Acad Sci U S A
2008;105:10513-8.[LinkOut]
- Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184
as Potential Oncogenic microRNA of Squamous Cell Carcinoma of
Tongue. Clin Cancer Res 2008;14:2588-92.[LinkOut]
- Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, et al.
A robust methodology to study urine microRNA as tumor marker:
microRNA-126 and microRNA-182 are related to urinary bladder
cancer. Urol Oncol 2010;28:655-61.[LinkOut]
- Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, et al. Differential
expression of microRNAs in plasma of patients with colorectal cancer: a
potential marker for colorectal cancer screening. Gut 2009;58:1375-81.[LinkOut]
- Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs
are promising novel biomarkers for early detection of colorectal cancer.
Int J Cancer 2010;127:118-26.[LinkOut]
- Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, Boland CR, et al.
Fecal MicroRNAs as novel biomarkers for colon cancer screening.
Cancer Epidemiol Biomarkers Prev 2010;19:1766-74.[LinkOut]
- Yamao T, Matsumura Y, Shimada Y, Moriya Y, Sugihara K, Akasu T,
et al. Abnormal expression of CD44 variants in the exfoliated cells
in the feces of patients with colorectal cancer. Gastroenterology
1998;114:1196-205.[LinkOut]
- Matsushita H, Matsumura Y, Moriya Y, Akasu T, Fujita S, Yamamoto S,
et al. A new method for isolating colonocytes from naturally evacuated
feces and its clinical application to colorectal cancer diagnosis.
Gastroenterology 2005;129:1918-27.[LinkOut]
- Onouchi S, Matsushita H, Moriya Y, Akasu T, Fujita S, Yamamoto S, et
al. New method for colorectal cancer diagnosis based on SSCP analysis
of DNA from exfoliated colonocytes in naturally evacuated feces.
Anticancer Res 2008;28:145-50.[LinkOut]
- Koga Y, Yasunaga M, Moriya Y, Akasu T, Fujita S, Yamamoto S, et
al. Detection of colorectal cancer cells from feces using quantitative
rea l-time RT-PCR for colorectal cancer diagnosis. Cancer Sci
2008;99:1977-83.[LinkOut]
- Koga Y, Yasunaga M, Takahashi A, Kuroda J, Moriya Y, Akasu T, et
al. MicroRNA expression profiling of exfoliated colonocytes isolated
from feces for colorectal cancer screening. Cancer Prev Res (Phila)
2010;3:1435-42.[LinkOut]
- Koga Y, Yasunaga M, Katayose S, Moriya Y, Akasu T, Fujita S, et
al. Improved recovery of exfoliated colonocytes from feces using
newly developed immunomagnetic beads.Gastroenterol Res Pract
2008;2008:605273.[LinkOut]
- Kanaoka S, Yoshida K, Miura N, Sugimura H, Kajimura M. Potential usefulness of detecting cyclooxygenase 2 messenger RNA in feces for
colorectal cancer screening. Gastroenterology 2004;127:422-7.[LinkOut]
- Takai T, Kanaoka S, Yoshida K, Hamaya Y, Ikuma M, Miura N, et
al. Fecal cyclooxygenase 2 plus matrix metalloproteinase 7 mRNA
assays as a marker for colorectal cancer screening. Cancer Epidemiol
Biomarkers Prev 2009;18:1888-93.[LinkOut]
- Hood JL, Pan H, Lanza GM, Wickline SA. Paracrine induction of
endothelium by tumor exosomes. Lab Invest 2009;89:1317-28.[LinkOut]
- Luo SS, Ishibashi O, Ishikawa G, Ishikawa T, Katayama A, Mishima T,
et al. Human villous trophoblasts express and secrete placenta-specific
microRNAs into maternal circulation via exosomes. Biol Reprod
2009;81:717-29.[LinkOut]
- Chang KH, Mestdagh P, Vandesompele J, Kerin MJ, Miller N.
MicroRNA expression profiling to identify and validate reference genes for relative quantification in colorectal cancer. BMC Cancer
2010;10:173.[LinkOut]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T.
MicroRNA-21 regulates expression of the PTEN tumor suppressor gene
in human hepatocellular cancer. Gastroenterology 2007;133:647-58.[LinkOut]
- Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post
S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates
tumor suppressor Pdcd4 and stimulates invasion, intravasation and
metastasis in colorectal cancer. Oncogene 2008;27:2128-36.[LinkOut]
- Hamaya Y, Yoshida K, Takai T, Ikuma M, Hishida A, Kanaoka S.
Factors that contribute to faecal cyclooxygenase-2 mRNA expression in
subjects with colorectal cancer. Br J Cancer 2010;102:916-21.[LinkOut]
Cite this article as:
Koga Y, Yasunaga M, Moriya Y, Akasu T, Fujita S, Yamamoto S, Matsumura Y. Exosome can prevent RNase from degrading microRNA in
feces. J Gastrointest Oncol. 2011;2(4):215-222. DOI: 10.3978/j.issn.2078-6891.2011.015
|