Study of cofilin 1 gene expression in colorectal cancer
Introduction
Colorectal cancer is the third and the fourth most prevalent cancer in women and men respectively and the fourth common cause of cancer death worldwide (1-3). This cancer is one of the most common malignancies with over 1 million cases diagnosed and 700,000 deaths per year worldwide (1,3). Five-year survival rate among people in North America, Eastern Europe and Iran is 65%, 54% and 43% respectively with improved survival if detected at an earlier stage (3,4). Therefore, detection of useful biomarkers with high sensitivity and specificity is a key step in diagnosis and target therapies for colorectal cancer (5,6).
Cofilin belongs to the actin depolymerization factor ADF/cofilin family (7), consisting of two muscle (CFL2) and non-muscle (CFL1) isoforms. CFL1 is a small ubiquitin protein with low molecular weight (~19 KDa) that involves in various functions in normal cells including cytokinesis, endocytosis, apoptosis and cell migration (8-13). Previous evidences suggest that cofilin 1 plays a major role in cell mobility. It is also reported that this protein is essential for invasion and metastasis of various human malignant solid tumors (9,10,14).
Tumor cell migration is essential for metastasis ability that is mediated by actin cytoskeleton (15). CFL1 is one of the key regulatory proteins in this process that is involved in actin filaments polymerization and depolymerization in a PH-dependent manner during cell migration (15,16).
Overexpression of CFL1 has been reported in some malignancies including prostate cancer, lung cancer, breast cancer and ovarian cancer (8,17-20) which is generally associated with metastasis.
In present study, we analyzed CFL1 expression levels in colorectal cancer tissues in comparison with adjacent non-tumor tissues, and investigated the diagnostic and prognostic value of this protein in colorectal cancer.
Methods
Patient samples
Thirty pairs of colorectal cancerous and non-cancerous (≥4 cm away from tumor margins) biopsies were obtained from patients who had undergone curative resection at the Surgical Department of Imam-Reza Hospital (Tabriz, Iran) without prior chemotherapy or radiotherapy from 2013 to 2016. Specimens were transmitted to RNase free micro-tubes and were snap-frozen in liquid nitrogen before transferring to −80 °C freezers. Demographic and clinicopathological data including age, gender, location of tumor, tumor stage and smoking were recorded and TNM staging was performed according to American Joint Committee on Cancer (AJCC).
The Medical Ethics Committee of Imam-Reza Hospital approved the study and all patients provided written informed consent for the study (medical ethics number: tbzmed.irec.1394.517).
RNA preparation
Total RNA was extracted from colorectal tumor tissues and adjacent non-tumor tissues using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. RNA quality was assessed using Picodrop Spectrophotometer (Picodrop Ltd., Hinxton, UK).
To remove any probable DNA remained from RNA extraction procedure, RNA was treated with DNase I enzyme. In which 10 µL reaction contained 2 µg RNA, 1 µL (1 U) DNase I enzyme, 1 µL DNase I buffer (10×), 0.5 µL RNase inhibitor enzyme and DEPC-treated water. The mixture was incubated at 37 °C for 30 min. After completion of the reaction, to inactivate DNase I enzyme, 1 µL of EDTA was added to the solution and incubated at 65 °C for 10 min.
cDNA synthesis
Reverse transcription polymerase chain reaction (RT-PCR) was performed to synthesize cDNA from prepared RNA. One µL dNTP mix (10 mM), 0.5 µL RT enzyme, 2 µL RT reaction buffer (5×), 0.5 µL oligo dT and 5.5 µL of RNA was incubated at 42 °C for 60 min. To inactivate the RT enzyme the solution was incubated at 85 °C for 5 minutes.
Real-time PCR
Light Cycler® 96 Real-Time PCR (Roche Molecular Systems, Inc., Pleasanton, CA, USA) system was applied to quantify CFL1 quantity in final cDNA product using SYBR Green PCR Master Mix. Each reaction solution contained 1 µL mix of specific forward and reverse primers, 10 µL SYBER Green Real-Time PCR Master Mix, 8 µL ddH2O and 1 µL reverse transcription product. Amplification condition was as follows: initial denaturation at 94 °C for 30 seconds followed by 40 cycles of denaturation at 94 °C for 10 seconds, annealing at 58 °C for 20 seconds and extension at 72 °C for 20 seconds followed by a final elongation step for 2 min at 72 °C. Primer sequences were as follows: CFL1 forward: 5'-TCTCGTCTTCTGCGGCTCTC-3'; CFL1 reverse: 5'-TCCAGGATGATGTTCTTCTTGTC-3'. Each sample was examined in triplicate and all results were normalized relative to glyceraldehyde phosphodehydrogenase (GAPDH) corresponding Cts. The quantified concentrations were reported as Ct values. The cycle number at which the generated fluorescence within a reaction crosses the threshold is considered as threshold cycle or Ct. CFL1 relative amount was calculated using the equation 2−∆Ct where Ct values were detected by thermocycler as the cycle at which the fluorescence absorbed by system passed the threshold.
∆Ct = CtCFL1 − CtGAPDH
Statistical analysis
Statistical analyses were made using the SPSS 21 software (SPSS Inc.) and SigmaPlot 13 statistical software. All data were expressed as means ± standard deviation (SD). Differences in CFL1 expression levels between colorectal cancer tissues and adjacent non-tumor tissues were analyzed using t-test. The relationship between CFL1 expression and clinicopathological features was assessed using ANOVA and t-test for samples. Receiver operating characteristic (ROC) curve was used to measure the possibility of using CFL1 as a diagnostic biomarker for the detection of colorectal cancer. A P value less than 0.05 was designated for results to be statistically significant.
Results
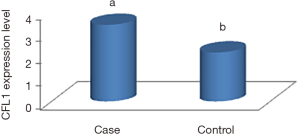
Using real-time RT-PCR, the expression levels of CFL1 in 30 pairs of samples of colorectal cancerous and non-cancerous tissues were analyzed. The analysis of results by SigmaPlot and SPSS softwares showed that expression of CFL1 was significantly higher in tumor tissue samples compared to their paired adjacent non-cancerous tissue samples (P<0.05) (Figure 1).
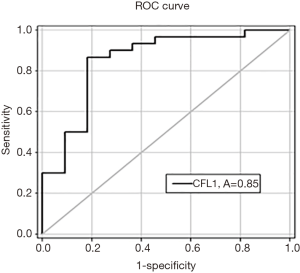
The relation between CFL1 expression and clinicopathological features such as age, sex, stage, smoking and location of tumors were analyzed as well (Table 1). The data showed no significant correlation between CFL1 expression levels and clinicopathological characteristics of patients. The analysis by SigmaPlot software to evaluate CFL1 capability as colorectal cancer tumor marker showed the area under ROC curve was 0.85 and the sensitivity and specificity were 82% and 97% respectively (Figure 2) indicating CFL1’s potential as biomarker for this malignancy.
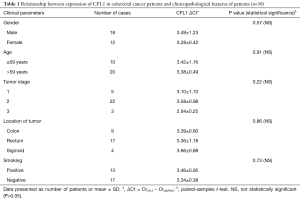
Full table
Discussion
Cofilin 1 is one of the important proteins responsible for cell migration process and plays a key role in the dynamics of actin filaments. Up-regulated actin cytoskeleton enhances tumor cell migration and invasion (21,22). Increased cofilin 1 expression has been reported in some malignancies including prostate, lung, breast and ovarian cancers (8,17-20) raising the possibility of CFL1 as a new molecular marker for some cancers.
Since colorectal cancer is the fourth common cause of cancer death worldwide (1-3), this detection would help early diagnosis of patients and might lead to better curative results. Involvement of CFL1 in cell migration and reported up-regulation of CFL1 in the mentioned cancers candidates this gene as a potential molecular biomarker for colorectal cancer. However, our survey in data resources showed no study concerning CFL1’s correlation to colorectal cancer.
In the present study, using real-time PCR technique, the expression of CFL1 was investigated in 30 patients with colorectal cancer. The results demonstrated that the expression of cofilin 1 was significantly higher in colorectal cancerous tissue samples compared to their paired adjacent non-cancerous tissue samples. Moreover, the relation between CFL1 expression and clinicopathological features of colorectal cancer patients was analyzed. The results proved no correlation between CFL1 expression and clinicopathological parameters of patients. The findings were in agreement with previous studies. Wang et al. (5) reported that the expression of CFL1 is significantly higher in pancreatic tumor tissues compared to noncancerous tissues while cofilin-2 expression was clearly reduced in cancerous tissues. Therefore, cofilin isoforms may serve as a clinical biomarker for pancreatic cancer patients. In an experiment, Collazo et al. (23) demonstrated that the level of active CFL1 increased in mouse and human models of prostate tumor metastasis, however this upregulation was enhanced in metastatic samples. In another study, Zheng et al. (18) analyzed serum content of cofilin protein using enzyme-linked immunosorbent assay in patients with different stages of lung cancer and indicated that the level of serum cofilin increases in patients with lung cancer, especially in cases with advanced stages. Furthermore, Zhou et al. (17) showed that overexpression of cofilin 1 may result in increased progression of ovarian carcinoma. They demonstrated that targeting the activities in tumor cells adequately prevents invasiveness of cancer cells.
CFL1 serves actin filaments and regulates actin polymerization and depolymerization during cell migration (24,25). Moreover, Yamaguchi et al. and Wang et al. (14,21) demonstrated cofilin’s active role in tumor invasiveness and metastasis. In a study by Yamaguchi et al. (26), it was found that suppression of cofilin 1 activity with small interfering RNA reduces invasiveness of carcinoma cells by reducing the assembly and stability of invadopodia. Another study by Lu et al. (8,20) showed that cofilin 1 may specifically help to predict prostate cancer progression and CFL1 expression in prostate mesenchyme may be closely related to the tumor progression and metastasis to lymph nodes. Another study done by Steller et al. (27) demonstrated that CFL1 activation is essential for the formation of membrane protrusions through CD95 death receptor and increased tumor cell invasion. Also, Zhang and Tong (28) reported cofilin 1 upregulation in samples of breast cancer with stages T0–T1 and T2 and suggested CFL1 as potential target for cancer therapy.
In the present study, we evaluated the diagnostic value of CFL1 as colorectal cancer tumor marker and found the area under ROC curve was high (0.85) with the sensitivity and specificity of 85% and 97% respectively (Figure 2).
In conclusion, the study confirmed that overexpression of CFL1 in samples of colorectal cancerous tissues compared to non-cancerous tissues, may play an important role as an oncogene in carcinogenesis of colorectal cancer. The findings support cofilin 1 as a novel diagnostic indicator in patients with colorectal cancer and suggests CFL1’s capability as a potent molecular target in colorectal cancer medication.
Acknowledgements
The authors are grateful to the Immunology Research Center, Tabriz University of Medical Sciences, Dr. Behzad Baradaran and Milad Asadi who sincerely helped us in this project.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The Medical Ethics Committee of Imam-Reza Hospital approved the study and all patients provided written informed consent (medical ethics number: tbzmed.irec.1394.517).
References
- Ling Y, Yang L, Huang H, et al. Prognostic significance of statin use in colorectal cancer: a systematic review and meta-analysis. Medicine (Baltimore) 2015;94. [Crossref] [PubMed]
- Pardini B, Naccarati A, Vodicka P, et al. Gene expression variations: potentialities of master regulator polymorphisms in colorectal cancer risk. Mutagenesis 2012;27:161-7. [Crossref] [PubMed]
- Shroff J, Thosani N, Batra S, et al. Reduced incidence and mortality from colorectal cancer with flexible-sigmoidoscopy screening: A meta-analysis. World J Gastroenterol 2014;20:18466-76. [Crossref] [PubMed]
- Moradi A, Khayamzadeh M, Guya M, et al. Survival of colorectal cancer in Iran. Asian Pac J Cancer Prev 2009;10:583-6. [PubMed]
- Wang Y, Kuramitsu Y, Ueno T, et al. Differential expression of up-regulated cofilin-1 and down-regulated cofilin-2 characteristic of pancreatic cancer tissues. Oncol Rep 2011;26:1595-9. [PubMed]
- Ghaffarzadehgan K, Jafarzadeh M, Raziee HR, et al. Expression of cell adhesion molecule CD44 in gastric adenocarcinoma and its prognostic importance. World J Gastroenterol 2008;14:6376-81. [Crossref] [PubMed]
- Bamburg JR, McGough A, Ono S. Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol 1999;9:364-70. [Crossref] [PubMed]
- Tsai CH, Lin LT, Wang CY, et al. Over-expression of cofilin-1 suppressed growth and invasion of cancer cells is associated with up-regulation of let-7 microRNA. Biochim Biophys Acta 2015;1852:851-61. [Crossref] [PubMed]
- Ghosh M, Song X, Mouneimne G, et al. Cofilin promotes actin polymerization and defines the direction of cell motility. Science 2004;304:743-6. [Crossref] [PubMed]
- Klamt F, Zdanov S, Levine RL, et al. Oxidant-induced apoptosis is mediated by oxidation of the actin-regulatory protein cofilin. Nat Cell Biol 2009;11:1241-6. [Crossref] [PubMed]
- Okreglak V, Drubin DG. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J Cell Biol 2007;178:1251-64. [Crossref] [PubMed]
- Kim YB, Choi S, Choi MC, et al. Cell adhesion-dependent cofilin serine 3 phosphorylation by the integrin-linked kinase.c-Src complex. J Biol Chem 2008;283:10089-96. [Crossref] [PubMed]
- Nagaoka R, Abe H, Kusano K, et al. Concentration of cofilin, a small actin-binding protein, at the cleavage furrow during cytokinesis. Cell Motil Cytoskeleton 1995;30:1-7. [Crossref] [PubMed]
- Wang W, Mouneimne G, Sidani M, et al. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J Cell Biol 2006;173:395-404. [Crossref] [PubMed]
- van Rheenen J, Condeelis J, Glogauer M. A common cofilin activity cycle in invasive tumor cells and inflammatory cells. J Cell Sci 2009;122:305-11. [Crossref] [PubMed]
- DesMarais V, Ghosh M, Eddy R, et al. Cofilin takes the lead. J Cell Sci 2005;118:19-26. [Crossref] [PubMed]
- Zhou J, Wang Y, Fei J, et al. Expression of cofilin 1 is positively correlated with the differentiation of human epithelial ovarian cancer. Oncol Lett 2012;4:1187-90. [Crossref] [PubMed]
- Zheng Y, Fang Y, Li S, et al. Detection of plasma cofilin protein for diagnosis of lung cancer. Nan Fang Yi Ke Da Xue Xue Bao 2013;33:1551-3. [PubMed]
- Li H, Zhang B, Liu Y, et al. EBP50 inhibits the migration and invasion of human breast cancer cells via LIMK/cofilin and the PI3K/Akt/mTOR/MMP signaling pathway. Med Oncol 2014;31:162. [Crossref] [PubMed]
- Lu LI, Fu NI, Luo XU, et al. Overexpression of cofilin 1 in prostate cancer and the corresponding clinical implications. Oncol Lett 2015;9:2757-61. [Crossref] [PubMed]
- Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta 2007;1773:642-52. [Crossref] [PubMed]
- Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol 1999;15:185-230. [Crossref] [PubMed]
- Collazo J, Zhu B, Larkin S, et al. Cofilin drives cell-invasive and metastatic responses to TGF-β in prostate cancer. Cancer Res 2014;74:2362-73. [Crossref] [PubMed]
- Müller CB, de Barros RL, Castro MA, et al. Validation of cofilin-1 as a biomarker in non-small cell lung cancer: application of quantitative method in a retrospective cohort. J Cancer Res Clin Oncol 2011;137:1309-16. [Crossref] [PubMed]
- Yap CT, Simpson TI, Pratt T, et al. The motility of glioblastoma tumour cells is modulated by intracellular cofilin expression in a concentration-dependent manner. Cell Motil Cytoskeleton 2005;60:153-65. [Crossref] [PubMed]
- Yamaguchi H, Lorenz M, Kempiak S, et al. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J Cell Biol 2005;168:441-52. [Crossref] [PubMed]
- Steller EJ, Ritsma L, Raats DA, et al. The death receptor CD95 activates the cofilin pathway to stimulate tumour cell invasion. EMBO Rep 2011;12:931-7. [Crossref] [PubMed]
- Zhang Y, Tong X. Expression of the actin-binding proteins indicates that cofilin and fascin are related to breast tumour size. J Int Med Res 2010;38:1042-8. [Crossref] [PubMed]


