Beyond microsatellite testing: assessment of tumor mutational burden identifies subsets of colorectal cancer who may respond to immune checkpoint inhibition
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer globally with estimated 1.4 million new cases and 694,000 deaths in 2012 (1). Contemporary median survivals approaching 30 months in the metastatic setting are seen with fluoropyrimidine-based combinations (FOLFOX, FOLFIRI) (2,3). However, after failure of oxaliplatin and irinotecan containing regimens, prolonged survival is uncommon (4,5).
Immune checkpoint inhibitor response has been observed in a growing number of clinical indications (6). Outside of melanoma, responses are seen in 15–20% of patients treated with single agent PD-1 or PD-L1 blocking antibodies across anatomic tumor types (7-9). However, reliable biomarkers capable of predicting response are needed. Increased neo-antigenic burden within tumor cells has been linked to PD-1/PD-L1 therapeutic response in several indications, however the high cost and significant time associated with neo-antigen discovery/prediction necessitates a more clinically relevant means of predicting response (7,10-12). Microsatellite instability (MSI) status, a genomic signature characterized by deficiencies in the mismatch repair (MMR) proteins MLH1, MSH2, MSH6, and/or PMS2 and accumulation of short tandem repeating segments of DNA (microsatellites), has emerged as a surrogate for increased tumor mutational burden (TMB). The clinical utility of MSI screening is predicated on identification of microsatellites in the genome of tumor cells either through polymerase chain reaction (PCR), or via immunohistochemical (IHC) staining to determine MMR protein integrity (13,14). Clinical studies have established MSI status as a putative response biomarker for PD-1 blockade, with progression free survival (PFS) rates of up to 78% reported in MSI-high (MSI-H) colorectal patients, compared to only 11% of microsatellite stable (MSS) patients (11,15). However, the mechanism that drives therapeutic response, increased neo-antigen burden, is only partially characterized by MSI status alone. Recently, evaluation of TMB through next-generation sequencing based comprehensive genomic profiling (CGP) has demonstrated utility in replacing standard MSI screening in CRC patients, with the added benefit of providing additional relevant genomic findings in genes such as EGFR, KRAS, BRAF and NRAS (16,17). Tumor mutational burden derived from CGP may represent a more robust surrogate for predicting response to PD-1 blockade and can be derived from CGP data. Herein, we explore the feasibility and potential utility of calculating TMB from a next-generation sequencing based CGP panel as a potential predictive biomarker of PD1/PD-L1 therapy in CRC.
Methods
Formalin-fixed, paraffin embedded tissue sections from 6,004 cases of histologically confirmed CRC were collected from 1,178 unique sites and sequenced using a hybrid capture-based comprehensive genomic profiling (CGP) assay (FoundationOne) (18). Patient demographics were captured and annotated to CGP results, including MSI and TMB status. Approval for this study, including a waiver of informed consent and a HIPAA waiver of authorization, was obtained from the Western Institutional Review Board (Protocol No. 20152817).
MSI methods
To determine MSI status using sequencing data generated via a CGP protocol, 114 intronic homopolymer repeat loci with adequate coverage on the CGP panel are analyzed for length variability and compiled into an overall MSI score via principal components analysis (19). Ranges of the MSI score were assigned MSI-high (MSI-H), MSI-ambiguous, or microsatellite stable (MSS) by manual unsupervised clustering of specimens for which MSI status was previously assessed either via IHC if available or approximated by the number of homopolymer indel mutations detected by the FoundationOne assay. This method of determining MSI status was validated for accuracy against currently approved methods, including immunohistochemistry and polymerase chain reaction based assessments, with results demonstrating 95% sensitivity and 98% specificity (n=69). Furthermore, precision of comprehensive genomic profiling based MSI calling was evaluated across 86 replicates spanning MSI-High to MSS status, and determined to be 100% for all evaluated samples (manuscript under review) (19).
TMB methods
TMB was calculated by counting the number of synonymous and non-synonymous mutations across a 1.11 megabase (Mb) region spanning 315 genes, with computational germline status filtering, and reporting the result as mutations/Mb (mut/Mb). This method has been previously validated for accuracy against whole exome sequencing (20). Patients were classified as TMB-high (≥11.7 mut/Mb) according to a 90% confidence interval based upon a Weibull distribution of TMB values observed within the MSI-high subgroup. Precision of the TMB values was validated in a separate cohort of 49 patients, replicated 4–6 times each. The TMB value of each patient ranged from 1.8 to 52.2 mut/Mb in the validation cohort. Reproducibility of the TMB status was evaluated against the threshold of 11.7 mutations/Mb used to identify TMB-high for this cohort. Results from the reproducibility evaluation demonstrated that 47 of 49 samples (96%) maintained the same TMB status, and the average coefficient of variation for the TMB score (mut/Mb) was determined to be 15% across all samples (Table S1).
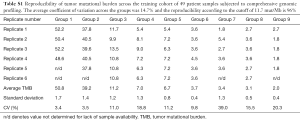
Full table
Statistical methods
In the TMB reproducibility study, the average TMB and coefficient of variation was determined from the replicate samples using statistical software provided by Microsoft Excel® (2016 MS Office). Statistical analysis comparing the frequency of somatic variants occurring in either the MSI-H or MSS groups, as well as TMB-high or TMB-low groups within the MSS cohort, was performed using a z-test calculated with JMP software (©SAS Institute, Inc.). Resulting P values were generated to determine significance. In order to identify a cutoff to classify TMB high patients, the distribution of TMB values across all 302 MSI-high patients was fit to a Weibull model, with a goodness of fit test achieving a P value <0.01 through a Cramer-von Mises W test using JMP software (©SAS Institute, Inc.). The TMB cutoff of ≥11.7 mutations/Mb equates to the lower bound of a 90% confidence interval of the expected TMB values associated with MSI-high according to a Weibull fit distribution.
Results
Patient characteristics
A total of 6,004 cases of CRC consisting of 2,817 (46.9%) women and 3,187 (53.1%) men were evaluated from the Foundation Core database between December 2014 and January 2017 across 1,178 medical centers. The median age at the time of tissue biopsy was 55.5 years (range, 8–88 years) in the overall population. Among cases deemed MSI-H, the median age at the time of biopsy was 63.0 years, with 148 female (49.0%) cases and 154 male (51.0%) cases. The median age within the MSS cases was slightly younger at 59 years, with 2,669 (46.8%) female cases and 3,033 (53.2%) male cases.
MSI status and tumor mutational burden
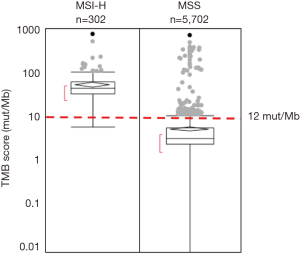
Overall, 5,702 cases (95.0%) were determined to be microsatellite stable (MSS) and 302 cases (5.0%) to be MSI-H by CGP analysis. The reported range of TMB within the total cohort (n=6,004) was 0 to 746.9 mutations/Mb (mut/Mb), with a median of 4.5 mut/Mb (Figure 1). The reported range of all MSI-H cases was 6.3–746.9 mut/Mb, compared to a range of 0–703.6 mut/Mb within the MSS cohort. The median TMB was significantly higher in the MSI-H compared to the MSS cohort (46.8 vs. 3.6 mut/Mb, P value <0.0001), consistent with the premise that loss of function in mismatch repair genes associated with MSI-H status contributes to a higher overall TMB. Nearly all (301/302, 99.7%) MSI-H patients were classified as TMB-high (≥11.7 mut/Mb), and the association of MSI-H status with high TMB was highly statistically significant (P<0.0001). Analysis of the 5,702 MSS CRC cases revealed that 164 (2.9%) were classified as TMB-high (range, 11.7–703.6 mut/Mb) (Figure 1).
Genomic alterations
To further investigate the genomic context of TMB-low/MSS, TMB-high/MSS and MSI-H samples we examined incidence of co-occurring known or likely oncogenic mutations in the genes NRAS, APC, TP53, PIK3CA, BRAF, KRAS and EGFR (Table 1). Additionally, we recorded the frequency of patients harboring a known or likely alteration in the MMR and DNA proof reading genes MLH1, MSH2, MHS6, PMS2, POLE and POLD1. Comparison between MSI-H and MSS cases, regardless of TMB score, demonstrated that BRAF, EGFR, PIK3CA or ERBB2 variants occurred more frequently in the MSI-H cohort, while variants in TP53, NRAS, KRAS, or APC were observed less frequently, respectively (Table 1). As anticipated, patient samples with at least one known or likely driver variant in the MMR genes MLH1, MSH6, MSH2 or PMS2 were highly enriched in the MSI-H population compared to the MSS population (18.5% vs. 0.2%; 28.1% vs. 0.5%, 15.6% vs. 0.4%, 5.3% vs. 0.2%, respectively; P value <0.0001). Known or likely driver events in the proofreading gene POLD1 were more frequently observed in the MSI-H cohort (1.3% vs. 0.1%, respectively; P value =0.0604). Variants within POLE were more frequently observed in the MSS cohort (0.8% vs. 0.3%, respectively; P value =0.1230) (Table 1).
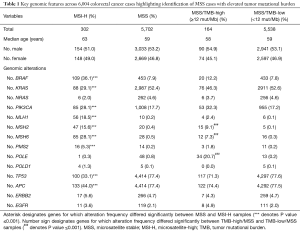
Full table
Evaluation of the TMB-high vs. TMB-low groups within the MSS cohort revealed that TMB-high/MSS patients were approximately 100× more likely to harbor known and likely variants in MSH2 (9.1% vs. 0.1%, respectively; P value <0.0001) and POLE (20.7% vs. 0.2%, P value <0.0001) compared to TMB-low/MSS patients. Among all of the POLE variants found in the TMB-high/MSS cohort, P286R, V411L and A456P accounted for 51% of the total. Variants in MSH6 (7.3% vs. 0.3%; P value =0.0537) and MLH1 (2.4% vs. 0.1%; P value <0.0001) were approximately 20× more frequent in TMB-high/MSS vs. TMB-low/MSS patients, and variants in PMS2 (1.8% vs. 0.2%; P value =0.1201) occurred about 9× more frequently in TMB-high/MSS vs. TMB-low/MSS patients. Variants in POLD1 were not observed with significant frequency (<0.1%) in either TMB-high or TMB-low patients within the MSS cohort. Other driver genes were found at less than 3× differences between the TMB-high/MSS and TMB-low/MSS groups (Table 1).
Illustrative case 1
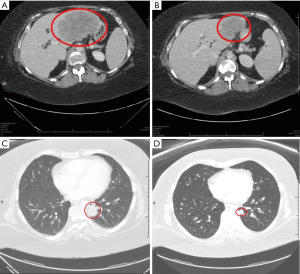
A 35-year-old female was diagnosed with stage IV rectal adenocarcinoma after presenting with rectal and buttock pain. Her tumor was KRAS wild type by hotspot PCR covering codons 12/13/61, MSS (by IHC), and germline negative for MLH1, MSH2, MSH6 and PMS2 alterations during testing for Lynch syndrome. She progressed on FOLFOX and panitumumab and second line FOLFIRI with worsening hepatic metastases and pelvic pain (Figure 2). To guide treatment options a biopsy was subjected for CGP (FoundationOne) that confirmed MSS CRC, but revealed a highly elevated TMB (223 mutations/Mb), and mutations in KRAS (at codon 146), MSH2, PIK3CA, PMS2, BRCA2, PIK3CA, POLE, and TP53. Given her refractoriness to chemotherapy, poor performance status, and emerging data supporting benefit of PD-1 blockade in TMB-high tumors, she was treated off-label with pembrolizumab. She had rapid symptomatic improvement and a significant radiographic response at 12 weeks and continues to benefit, now 7 months since starting therapy (Figure 2).
Illustrative case 2
A 45-year-old Caucasian female was diagnosed with stage III rectal cancer, MSS by initial immunohistochemical analysis. Following initial therapy, she developed pelvic sidewall and biopsy-proven anastomotic recurrence but declined salvage surgical options or chemoradiotherapy. To explore maximal options her original surgical sample was sent for CGP, revealing a MSS, RAS/RAF wild type tumor with high TMB of 14 mut/Mb. Known alterations in APC and TP53 were additionally identified. She was started on off-label nivolumab and achieved a complete response, now lasting over 18 months with no endoscopic evidence of tumor.
Discussion
Herein, we report analysis from a large cohort of over 6,000 colorectal cancer patients and describe a TMB threshold that identifies 99.7% of MSI-H patients, while capturing an additional 3% of MSS samples, increasing the potential treatment population by 54%. Our series suggests TMB and MSI can be reliably derived from CGP data, and can identify an additional cohort of patients (MSS/TMB-high) who may benefit from immune checkpoint inhibitors.
Immuno-oncology studies have highlighted several potential biomarkers of varying large scale clinical feasibility including PD-1/L1 IHC, tumor infiltrating lymphocytes (TILs), and elevated numbers of nonsynonymous mutations (21-25). Elevated in-silico predicted class I neoantigen load is emerging as a robust predictive response biomarker to checkpoint inhibitor therapies, but has been derived from whole-exome sequencing (WES), a time and cost intensive method not widely available (10). Our series identifies the TMB distribution across MSI-H colorectal cancers, but more importantly suggests that nearly 1 in every 33 MSS colorectal patients have an elevated mutational burden according to a classification based upon MSI status. While MSI-H tumors are seen in 12–22% of stage II and III CRC respectively, MSI-H is observed in only 3–5% of stage IV patients (26,27). Thus, among the roughly 50,000 CRC-related deaths per year in the US, CGP has the potential to identify 1,500 patients/year with MSS/TMB-high tumors (28).
Interestingly, the CGP-derived genomic features associated with the MSS/TMB-high cohort are suggestive of a mismatch repair defective state that likely induces increased TMB through spontaneous POLE loss of function (Table 1). The implication of an enriched POLE genotype within the MSS/TMB-high group supports the hypothesis that defects in both the MMR and DNA proofreading pathway can cause a hypermutated state, without necessarily giving rise to the short tandem repeat signature observed through classic MSI-H testing. Our first patient case harbored the classic genomic features associated with the MSS/TMB-high group (MSH2+, POLE+). Pathogenic POLE aberrations have been seen to identify patients responding to PD-1 inhibitors in endometrial cancers (29-31). Case 2 highlights the observation that perhaps even a slightly elevated TMB increases the likelihood of an immunogenic neo-epitope to drive an immune response, as in theory only a single neo-epitope may be needed.
Recently, CGP-derived TMB (by the same assay used here) was identified as an independent predictive response marker in trials evaluating various checkpoint inhibitors and combinations in bladder cancer, NSCLC and metastatic melanoma (7,32,33). Further work is needed to determine if increasing TMB has a linear relationship with probability of response to immunotherapies, but our work suggests CGP can reliably determine TMB over a dynamic range reflective of patients seen in clinical practice. Notably, additional MSS/TMB-high cases (n=2) with exceptional responses to checkpoint inhibitor therapy are recently described, adding further support to our findings (34,35). Beyond TMB, it is important to note that MSI determination by NGS is not considered standard, however, mounting published data suggests excellent agreement with standard IHC and PCR methods (manuscript under review) (18).
The rates of MSI-H samples (5.0%) in our series is in agreement with the frequencies recorded from clinical trials in advanced CRC, suggesting our cohort accurately reflects the advanced CRC landscape (26). Similarly, KRAS and BRAF frequencies support this, however, we acknowledge a potential selection bias as samples subjected to the CGP assay (FoundationOne) used in our series may have been previously tested by orthogonal means. Additionally, we cannot draw conclusions about the responsiveness to immune-mediated therapies of MSS/TMB-high patients on the lower end of the TMB spectrum based on anecdotal evidence alone. While it can be argued that our first case harbors genomic alterations known to be associated with checkpoint inhibitor response it is important to note that neither the POLE mutation or PD-L1 and PD-L2 gene amplification would have been detected by standard of care testing in CRC.
In the trial by Le and colleagues using traditional MSI testing (Promega MSI Assay, Promega Corporation) no responses were observed in MSS colorectal cancers (15). However, the sample size of patients with MMR-proficient tumors (defined by MSI negative) was only 18, and not likely to have included an MSS/TMB-high tumor given the 3% incidence rate. Prospective knowledge of MSS/TMB-high status may well have resulted in responses to anti-PD-1 as suggested herein. To our knowledge, our study represents the largest series in CRC investigating TMB using a validated CGP assay, and identifies a 3% frequency of high mutational burden in MSS tumors. Further prospective study of MSS/TMB-high patients is warranted.
Acknowledgements
The authors would like to acknowledge the Campbell family for their contribution to this study.
Funding: This work was supported in part by the National Cancer Institute at the National Institutes of Health (RO1 CA172302 to AF Hezel).
Footnote
Conflicts of Interest: DA Fabrizio, G Frampton, J Sun, K Gowen, M Kennedy, J Greenbowe, AB Schrock, JS Ross, PJ Stephens, SM Ali, and VA Miller are employees and hold equity in Foundation Medicine, Inc. DA Fabrizio holds equity in Juno Therapeutics and Seattle Genetics. SJ Klempner has received honoraria from Foundation Medicine, Inc. The other authors have no conflicts of interest to declare.
Ethical Statement: Approval for this study, including a waiver of informed consent and a HIPAA waiver of authorization, was obtained from the Western Institutional Review Board (Protocol No. 20152817).
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- O'Neil BH, Venook AP. Trying to Understand Differing Results of FIRE-3 and 80405:Does the First Treatment Matter More Than Others? J Clin Oncol 2015;33:3686-8. [Crossref] [PubMed]
- Heinemann V, von Weikersthal LF, Decker T, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol 2014;15:1065-75. [Crossref] [PubMed]
- Grothey A, Van Cutsem E, Sobrero A, et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet 2013;381:303-12. [Crossref] [PubMed]
- Mayer RJ, Van Cutsem E, Falcone A, et al. Randomized trial of TAS-102 for refractory metastatic colorectal cancer. N Engl J Med 2015;372:1909-19. [Crossref] [PubMed]
- Pico de Coana Y, Choudhury A, Kiessling R. Checkpoint blockade for cancer therapy: revitalizing a suppressed immune system. Trends Mol Med 2015;21:482-91. [Crossref] [PubMed]
- Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 2016;387:1909-20. [Crossref] [PubMed]
- Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540-50. [Crossref] [PubMed]
- Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455-65. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Dudley JC, Lin MT, Le DT, et al. Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin Cancer Res 2016;22:813-20. [Crossref] [PubMed]
- Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189-99. [Crossref] [PubMed]
- Bairwa NK, Saha A, Gochhait S, et al. Microsatellite instability: an indirect assay to detect defects in the cellular mismatch repair machinery. Methods Mol Biol 2014;1105:497-509. [Crossref] [PubMed]
- You JF, Buhard O, Ligtenberg MJ, et al. Tumours with loss of MSH6 expression are MSI-H when screened with a pentaplex of five mononucleotide repeats. Br J Cancer 2010;103:1840-5. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Salipante SJ, Scroggins SM, Hampel HL, et al. Microsatellite instability detection by next generation sequencing. Clin Chem 2014;60:1192-9. [Crossref] [PubMed]
- Stadler ZK, Battaglin F, Middha S, et al. Reliable Detection of Mismatch Repair Deficiency in Colorectal Cancers Using Mutational Load in Next-Generation Sequencing Panels. J Clin Oncol 2016;34:2141-7. [Crossref] [PubMed]
- Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023-31. [Crossref] [PubMed]
- Hall MJ, Gowen K, Sanford EM, et al. Evaluation of microsatellite instability (MSI) status in gastrointestinal (GI) tumor samples tested with comprehensive genomic profiling (CGP). J Clin Oncol 2016;34:abstr 528.
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [Crossref] [PubMed]
- McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463-9. [Crossref] [PubMed]
- Postow MA, Callahan MK, Wolchok JD. Immune Checkpoint Blockade in Cancer Therapy. J Clin Oncol 2015;33:1974-82. [Crossref] [PubMed]
- McLaughlin J, Han G, Schalper KA, et al. Quantitative Assessment of the Heterogeneity of PD-L1 Expression in Non-Small-Cell Lung Cancer. JAMA Oncol 2016;2:46-54. [Crossref] [PubMed]
- Bhaijee F, Anders RA. PD-L1 Expression as a Predictive Biomarker: Is Absence of Proof the Same as Proof of Absence? JAMA Oncol 2016;2:54-5. [Crossref] [PubMed]
- Schalper KA, Kaftan E, Herbst RS. Predictive Biomarkers for PD-1 Axis Therapies: The Hidden Treasure or a Call for Research. Clin Cancer Res 2016;22:2102-4. [Crossref] [PubMed]
- Koopman M, Kortman GA, Mekenkamp L, et al. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer 2009;100:266-73. [Crossref] [PubMed]
- Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer:results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol 2010;28:466-74. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29. [Crossref] [PubMed]
- Howitt BE, Shukla SA, Sholl LM, et al. Association of Polymerase e-Mutated and Microsatellite-Instable Endometrial Cancers With Neoantigen Load, Number of Tumor-Infiltrating Lymphocytes, and Expression of PD-1 and PD-L1. JAMA Oncol 2015;1:1319-23. [Crossref] [PubMed]
- Santin AD, Bellone S, Buza N, et al. Regression of chemotherapy-resistant Polymerase epsilon (POLE) ultra-mutated and MSH6 hyper-mutated endometrial tumors with nivolumab. Clin Cancer Res 2016;22:5682-7. [Crossref] [PubMed]
- Mehnert JM, Panda A, Zhong H, et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J Clin Invest 2016;126:2334-40. [Crossref] [PubMed]
- Johnson DB, Frampton GM, Rioth MJ, et al. Targeted Next Generation Sequencing Identifies Markers of Response to PD-1 Blockade. Cancer Immunol Res 2016;4:959-67. [Crossref] [PubMed]
- Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 2018. N Engl J Med 2018;378:2093-104. [Crossref] [PubMed]
- Gong J, Wang C, Lee PP, et al. Response to PD-1 Blockade in Microsatellite Stable Metastatic Colorectal Cancer Harboring a POLE Mutation. J Natl Compr Canc Netw 2017;15:142-7. [Crossref] [PubMed]
- Sorscher S, Desnoyers R. A Patient with A Microsatellite Stable (MSS) and High Mutational Burden Metastatic Colorectal Cancer Responding To Checkpoint Inhibitor Therapy. MOJ Clin Med Case Rep 2016;5:00135.


