KRAS, NRAS and BRAF analysis of ampullary adenocarcinoma classified using CK7, CK20, MUC1 and MUC2
Introduction
Ampullary carcinomas are rare and dominated by adenocarcinomas. They account for only 0.5% of all gastrointestinal malignancies (1,2) and induce 36% of all pancreaticoduodenectomies (3). Although having a better prognosis than pancreatic ductal adenocarcinoma, ampullary adenocarcinoma (AAC) remains a deadly disease, killing 60% of affected patients (4). AAC with pancreaticobiliary (PB) histology has a worse outcome than this with intestinal (IT) histology (2). The IT type originates from the IT epithelium overlying the ampulla and the PB type originates from the epithelium of the distal common bile duct, distal pancreatic duct, or common ampullary duct. In addition, mixed type (M) contains both epitheliums (5). This subclassification remains a challenge for pathologists and induces a reasonable level of disagreement. Immunohistochemistry (IHC) using cytokeratin (CK) and apomucin (MUC) could facilitate subtyping. In fact, PB type expresses CK7 and MUC1, while IT type expresses CK20 and MUC2 and M type expresses all these markers (6). Genetic features of these subtypes remain unclear. In this study, we aimed to evaluate differences in prognostic, pathological and molecular parameters including mutational status of three oncogenes KRAS, NRAS and BRAF, between these subtypes in AAC from 21 Tunisian patients; subtyped on HE and IHC using CK7, CK20, MUC1 and MUC2.
Methods
Cases selection
This retrospective study was approved by institutional ethics committee of Habib Thameur Hospital of Tunis (HTHEC-2017-03). Clinical, epidemiological and prognosis analysis; including the following parameters: age, sex, tumor size, TNM stage, differentiation, vascular emboli and perineural invasion, was determined referring to patients’ files. Formalin fixed paraffin embedded (FFPE) tissues, resected in the period 2000–2016, were obtained from the archival tissue collection of pathology department. All specimens were fixed in 10% buffered formalin. Cases were revised by two pathologists to subclassify tumors.
MUC and CK IHC
IHC labeling was carried out in an automated Leica Bond Max (Leica Microsystems, Germany) through the following mouse monoclonal antibodies (anti-CK7: NCL-L-CK7-560, clone RN7, 1:100; anti-CK20: mouse NCL-L-CK20-561, clone PW31, 1:100; anti-MUC1: NCL-MUC-1, clone Ma695, 1:100; and anti-MUC2: NCL-MUC-2, clone Ccp58, 1:100; Novocastra, Leica Biosystems, Newcastle, Upon, UK). Briefly, 3 µm thick sections were prepared from each bloc and were dried overnight at 60 °C. First, tissues were deparaffinized using xylene and pre-treated with the Epitope Retrieval Solution (EDTA-buffer, pH =8.8) in the rate of 98 °C for 20 min. After washing steps, peroxidase blocking was carried out for 10 min using the Bond Polymer Refine Detection Kit DC9800 (Leica Microsystems). Tissues were again washed and then incubated with primary antibody for 30 min at 25 °C. Subsequently, tissues were incubated with polymer for 10 min and developed with DAB-Chromogen for 10 min. IHC reaction was considered to be positive regardless the number of cells stained.
Histomolecular classification
Pancreatobiliary subtype
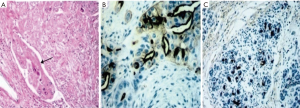
Most PB subtype have flat or micropapillary pattern, often complex with cribriform areas. The tumor cells are cubic without pseudostratification. Nuclei are enlarged and vesicular with dispersed chromatin. They possess varying degrees of mucosecretion with morphology similar to pancreatic ductal adenocarcinoma. In IHC, tumor cells express MUC1 and CK7.
IT subtype
Superimposed to digestive tract segments, the architecture of the lesion was tubulous, tubulo-villous or villous. Neoplastic cells are cylindrical with pseudostratified and elongated nuclei showing varying degrees of atypia. In IHC, tumor cells express CK20 and MUC2.
Mixed subtype
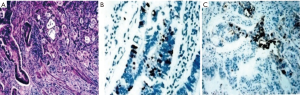
This subtype contains >10% of the two epitheliums, and express CK7, CK20, MUC1 and MUC2.
Molecular analysis
Molecular analysis was performed as described in (7). Briefly, the most representative blocks were selected. For each sample, 10 sections were used for DNA extraction. Genomic DNA extraction was performed according to Kit (QIAamp® DNA FFPE Tissue QIAGEN, Hilden, Germany) manufacturer’s handbook. Nine hotspot sites of KRAS, NRAS and BRAF were analyzed in this order—KRAS: codons 12 and 13, codons 59 and 61, codon 117 and codon 146, NRAS: codons 12 and 13, codons 59 and 61, codon 117 and codon 146 and BRAF: codon 600. After each pyrosequencing, the mutated samples were excluded and only wild type samples could be amplified for following sequencing. PCR was performed in 30 µL final volume with 2 U of Taq polymerase (500 U Taq polymerase, Agilent Technologies, Wilmington, USA), 1X PCR buffer, 0.1 mM dNTPs, 30 pM of each primer and 50 ng of genomic DNA. A wild-type genomic DNA (QIAGEN) was used as a negative control. Real time sequencing was performed using PyroMark Q24 pyrosequencing instrument and software according to the manufacturer’s instructions (www.pyrosequencing.com). Detailed information about methods and lists of primers’ sequences, sequences to analyze and dispensation orders are described in our previous study (7), and are listed in the following articles (8,9).
Statistical analysis
Data were processed using SPSS 20.0 statistical software (SPSS, Inc., USA). Patients’ characteristics were analyzed using descriptive statistics. Qualitative and quantitative variables were analyzed, as appropriate, using non-parametric tests, with a significant P value less than 0.05.
Results
Clinicopathological characteristics
Thirteen patients were women and 8 were men. Mean age at operation was 60 years (35–75 years). Mean tumor size was 2.2 cm (0.5–7.5 cm). Resection margins were positive in one case. Tumor was well differentiated [13], moderately differentiated [7] and poorly differentiated [1]. T1 stage was observed in 4 cases, T2 in 12 cases and T3 stage in 5 cases with N1 stage in 5 cases. Perineural invasion and vascular emboli were present in 2 and 4 cases, respectively. Associated lesions were PanIN in 10 cases. Perineural invasion was statistically associated with vascular emboli and differentiation degree (P=0.029 and 0.033, respectively). T stage was associated with tumor size and perineural invasion (P=0.049 and 0.056, respectively). Sex and N stage were statistically associated (P=0.047). CK7, CK20, MUC1 and MUC2 were positive in 17, 8, 18 and 7 cases, respectively. MUC1 influenced T stage (P=0.033) while N stage was statistically associated with MUC2 and CK20 (P=0.025 and 0.047, respectively). These data are shown in Table 1.
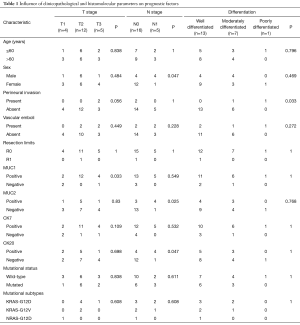
Full table
Histomolecular classification
Based on HE and IHC evaluation, three subtypes were obtained:
- Fifteen cases MUC1+/CK7+ were classified as PB (Figure 1);
- Two cases MUC2+/CK20+ were classified as IT (data not shown);
- Four cases MUC1+/MUC2+/CK7+/CK20+ were classified as M (Figure 2).
Genetic characteristics
Nine cases were mutated and 12 were wild-type. Eight cases were KRAS mutated (5 G12D and 3 G12V). Only one case was NRAS mutated (G12D). No BRAF mutation was found. Seven of the 15 PB cases and 2 of the 4 M cases were mutated. The two IT subtypes were wild-type. Mutations classes’ distribution through histomolecular subtypes and their influence on prognostic factors are shown in Table 1.
Histomolecular subtypes characteristics
Different characteristics of histomolecular subtypes are shown in Table 2. Briefly, 4/4 M and 1/15 PB subtype were N1, while 2/2 IT subtype, were N0. This difference was significant (P=0.001). Histomolecular subtype was correlated too with associated lesions (P=0.027). The only R1 case was PB subtype. The two IT subtype cases were wild-type, while 2/4 of M subtype and 7/15 of PB subtype were mutated.
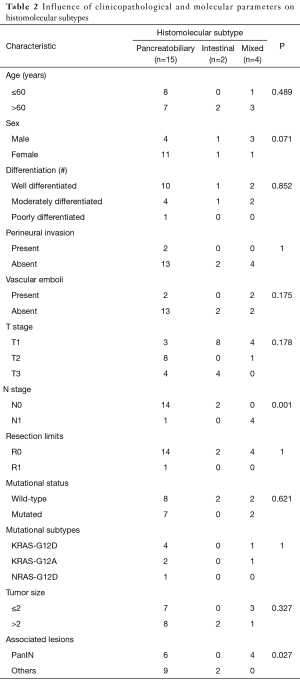
Full table
Discussion
In this study, we found a statistically significant difference between histological subtypes of AAC in N stage and precursor lesions. Histologically, the IT type evolves through adenoma-carcinoma sequences and the PB type arises from precursor large-duct PanIN (5). Concerning our cases, 15/21 were PB, 2 were IT and 4 were M; which is comparable to other studies’ findings. Indeed, the overall prevalence of the IT type ranges from 26% to 74%, the PB type from 22% to 72% and the M from 7% to 39% (4,6,10,11-24).
We found that IHC parameters (MUC1, CK7 and CK20) influence significantly T and N stage. In other studies, MUC1 and MUC2 expression was significantly associated with tumor differentiation, lymphatic and perineural invasion, tumor stage and survival (5,17).
Histomolecular subtypes are thought to be one of the most important prognostic factors for AAC. Patients with PB type have a worse overall survival than those with IT type (2,5,17-19,22). In addition, histomolecular subtypes are significantly associated with pathological grade, lymphatic and perineural invasion, tumor stage, CK7, CK20, MUC1 and MUC2 expression (4,5,11,13).
More recent studies have investigated additional markers such as DNA mutations to identify prognostically distinct AAC subtypes. Analysis revealed 19,143 genome-wide somatic point mutations, of which 30 maps within known annotated coding sequences. The most notable alteration is an activating KRAS mutation at codon 12 (G12V) (25). KRAS mutations were detected in 8/21 of our cases as other studies which found them in 23% to 47% of cases (4,5,11,12,19,26,27). Also, we found mutations in NRAS in one case. BRAF wasn’t mutated. Seven of the 15 PB and 2 of the 4 M were mutated. The 2 IT subtypes were wild-type. This prevalence is also in line with observations made by others. KRAS is mutated in 36% to 61% of PB subtype, in 5% to 52% of IT subtype and in 7% to 45% of M subtype. No mutation is detected in NRAS and BRAF is mutated in 0% to 9% of PB subtype, in 0% to 1% of IT subtype and in 0% to 1% of M subtype (4,5,14,19,28).
Studies analyzing the prognostic value of KRAS mutations in AAC reported conflicting results. In fact, KRAS mutations especially KRAS G12D are associated with shorter survival in some series (4,12,29,30). Interestingly, patients with mutations other than KRAS G12D do not appear to be different from those with KRAS wild-type (12). While, other groups don’t show any influence of KRAS or BRAF status on survival (18,19,30).
Only few studies have described the prognosis and characteristics of M subtype. M subtype was reported to have an intermediate prognosis between the PB and IT phenotypes (17). In the other side, others indicated that the pathological characteristics of the M subtype is similar to those of the PB subtype, and prognosis of the M subtype is poor in comparison to that of the IT phenotype (5). In our series, we couldn’t give more clarifications because of the limited number of M subtypes.
Our study has several limitations. First, our series was small. Then, our methodology was limited to DNA-level alterations without specific assessment of possible epigenetic mechanisms. It has recently been shown that differences in phenotypic differentiation of AAC are reflected in RNA profiling (15). Future trials should be designed in view of our increased understanding of the different anatomic and histomolecular profiled subtypes of these cancers.
In conclusion, we validate the prognostic utility of the histomolecular classification of AAC. This combination of HE and IHC classification should be incorporated into our clinical practice. We hypothesize that these tumors necessitate highly patient- and tumor-individualized therapy. Future efforts to understand the biological bases of these subgroups are needed.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical statement: The study was approved by the Habib Thameur Hospital Ethics Committee HTHEC (No. HTHEC-2017-03). As this was a retrospective study, informed consent is not required.
References
- Albores-Saavedra J, Schwartz AM, Batich K, et al. Cancers of the ampulla of vater: demographics, morphology, and survival based on 5,625 cases from the SEER program. J Surg Oncol 2009;100:598-605. [Crossref] [PubMed]
- Ahn DH, Bekaii-Saab T. Ampullary cancer: an overview. Am Soc Clin Oncol Educ Book 2014.112-5. [Crossref] [PubMed]
- Saavedra JA. WHO classification of tumors of the digestive system. IARC, Lyon 2010;31:27-31.
- Mafficini A, Amato E, Cataldo I, et al. Ampulla of Vater Carcinoma: Sequencing Analysis Identifies TP53 Status as a Novel Independent Prognostic Factor and Potentially Actionable ERBB, PI3K, and WNT Pathways Gene Mutations. Ann Surg 2018;267:149-56. [Crossref] [PubMed]
- Asano E, Okano K, Oshima M, et al. Phenotypic characterization and clinical outcome in ampullary adenocarcinoma. J Surg Oncol 2016;114:119-27. [Crossref] [PubMed]
- Kawabata Y, Tanaka T, Nishisaka T, et al. Cytokeratin 20 (CK20) and apomucin 1 (MUC1) expression in ampullary carcinoma: Correlation with tumor progression and prognosis. Diagn Pathol 2010;5:75. [Crossref] [PubMed]
- Ferchichi M, Jouini R, Ayari I, et al. Kras, epidermal growth factor receptor, and p53, but not Nras or Braf, biomarkers are frequently altered in pancreatic adenocarcinoma and precursor lesions. Drug Invention Today 2017;9:88-96.
- Jung A, Straße T. RAS mutation detection handbook, detection of mutations in exons 2, 3, and 4 of the KRAS and NRAS oncogenes. SOP collection provided by the RAS-Advisory Board of the DGP (German Society for Pathology) and BDP (Federation of German Pathologists), 2013.
- Spittle C, Ward MR, Nathanson KL, et al. Application of a BRAF pyrosequencing assay for mutation detection and copy number analysis in malignant melanoma. J Mol Diagn 2007;9:464-71. [Crossref] [PubMed]
- Tan J, Tan P, Teh BT. Defining the Molecular Alterations of Ampullary Carcinoma. Cancer Cell 2016;29:135-6. [Crossref] [PubMed]
- Kohler I, Jacob D, Budzies J, et al. Phenotypic and genotypic characterization of carcinomas of the papilla of Vater has prognostic and putative therapeutic implications. Am J Clin Pathol 2011;135:202-11. [Crossref] [PubMed]
- Valsangkar NP, Ingkakul T, Correa-Gallego C, et al. Survival in ampullary cancer: potential role of different KRAS mutations. Surgery 2015;157:260-8. [Crossref] [PubMed]
- Schueneman A, Goggins M, Ensor J, et al. Validation of histomolecular classification utilizing histological subtype, MUC1, and CDX2 for prognostication of resected ampullary adenocarcinoma. Br J Cancer 2015;113:64-8. [Crossref] [PubMed]
- Hechtman JF, Liu W, Sadowska J, et al. Sequencing of 279 cancer genes in ampullary carcinoma reveals trends relating to histologic subtypes and frequent amplification and overexpression of ERBB2 (HER2). Mod Pathol 2015;28:1123-9. [Crossref] [PubMed]
- Schultz NA, Werner J, Willenbrock H, et al. MicroRNA expression profiles associated with pancreatic adenocarcinoma and ampullary adenocarcinoma. Mod Pathol 2012;25:1609-22. [Crossref] [PubMed]
- Leo JM, Kalloger SE, Peixoto RD, et al. Immunophenotyping of ampullary carcinomata allows for stratification of treatment specific subgroups. J Clin Pathol 2016;69:431-9. [Crossref] [PubMed]
- Chang DK, Jamieson NB, Johns AL, et al. Histomolecular phenotypes and outcome in adenocarcinoma of the ampulla of vater. J Clin Oncol 2013;31:1348-56. [Crossref] [PubMed]
- Overman MJ, Zhang J, Kopetz S, et al. Gene expression profiling of ampullary carcinomas classifies ampullary carcinomas into biliary-like and intestinal-like subtypes that are prognostic of outcome. PLoS One 2013;8. [Crossref] [PubMed]
- Mikhitarian K, Pollen M, Zhao Z, et al. Epidermal growth factor receptor signaling pathway is frequently altered in ampullary carcinoma at protein and genetic levels. Mod Pathol 2014;27:665-74. [Crossref] [PubMed]
- Lau SK, Weiss LM, Chu PG. Differential expression of MUC1, MUC2, and MUC5AC in carcinomas of various sites: an immunohistochemical study. Am J Clin Pathol 2004;122:61-9. [Crossref] [PubMed]
- Perysinakis I, Minaidou E, Leontara V, et al. Differential Expression of β-Catenin, EGFR, CK7, CK20, MUC1, MUC2, and CDX2 in Intestinal and Pancreatobiliary-Type Ampullary Carcinomas. Int J Surg Pathol 2017;25:31-40. [Crossref] [PubMed]
- Morini S, Perrone G, Borzomati D, et al. Carcinoma of the ampulla of Vater: morphological and immunophenotypical classification predicts overall survival. Pancreas 2013;42:60-6. [Crossref] [PubMed]
- Perysinakis I, Margaris I, Kouraklis G. Ampullary cancer--a separate clinical entity? Histopathology 2014;64:759-68. [Crossref] [PubMed]
- Chandrasegaram MD, Chen JW, Price TJ, et al. Advances in Molecular Pathology and Treatment of Periampullary Cancers. Pancreas 2016;45:32-9. [Crossref] [PubMed]
- Demeure MJ, Craig DW, Sinari S, et al. Cancer of the ampulla of Vater: analysis of the whole genome sequence exposes a potential therapeutic vulnerability. Genome Med 2012;4:56. [Crossref] [PubMed]
- Moore PS, Orlandini S, Zamboni G, et al. Pancreatic tumours: molecular pathways implicated in ductal cancer are involved in ampullary but not in exocrine nonductal or endocrine tumorigenesis. Br J Cancer 2001;84:253-62. [Crossref] [PubMed]
- Oliveira-Cunha M, Hadfield KD, Siriwardena AK, et al. EGFR and KRAS mutational analysis and their correlation to survival in pancreatic and periampullary cancer. Pancreas 2012;41:428-34. [Crossref] [PubMed]
- Schönleben F, Qiu W, Allendorf JD, et al. Molecular analysis of PIK3CA, BRAF, and RAS oncogenes in periampullary and ampullary adenomas and carcinomas. J Gastrointest Surg 2009;13:1510-6. [Crossref] [PubMed]
- Schultz NA, Roslind A, Christensen IJ, et al. Frequencies and prognostic role of KRAS and BRAF mutations in patients with localized pancreatic and ampullary adenocarcinomas. Pancreas 2012;41:759-66. [PubMed]
- Kim BJ, Jang HJ, Kim JH, et al. KRAS mutation as a prognostic factor in ampullary adenocarcinoma: a meta-analysis and review. Oncotarget 2016;7:58001-6. [PubMed]


