Germline pharmacogenomics of DPYD*9A (c.85T>C) variant in patients with gastrointestinal malignancies treated with fluoropyrimidines
Introduction
Fluoropyrimidines includes intravenous fluorouracil (FU), its oral pre-prodrug capecitabine and the oral prodrug tegafur [component of tegafur-uracil and Teysuno (S1)] (1,2). Fluoropyrimidines are considered the cornerstone of most chemotherapeutic regimens approved for the treatment of gastrointestinal (GI) tract tumors. They are also commonly used in the treatment of other types of solid malignancies including breast and head and neck cancers (3,4).
Dihydropyrimidine dehydrogenase (DPD) is an enzyme (EC 1.3.1.2) encoded by DPYD gene. DPD is the rate-limiting enzyme for catabolism of uracil, thymine and their analogue fluoropyrimidine and eliminates >80% of administered or formed FU (5). Generally, treatment of cancer patients with fluoropyrimidines is relatively well tolerated. However, around 5–10% of the treated patients develop severe, potentially life threatening toxicity such as GI toxicity, skin toxicity, myelosuppression and neurotoxicity (6-8). Among patients with DPD deficiency, the incidence of grade 3 or greater toxicities has been reported to be as high as 88%. In those patients, fluoropyrimidines can be fatal (9-12). Moreover, toxicity develops significantly earlier in patients with low DPD activity than in patients with normal DPD activity (13).
The prevalence of DPD deficiency in Caucasian is approximately 3–5% (14). Profound deficiency of DPD is less frequent occurring in approximately 0.2% of individuals (15-20). Molecular analysis of patients with DPD deficiency has identified over 128 mutations and polymorphisms in the DPYD gene that may result in partial or total loss of DPD activity (17,21,22). Three DPYD variants (DPYD*2A, DPYD*13 and DPYD*9B) have consistently been reported to be associated with fluoropyrimidines-associated toxicity and impaired DPD enzyme activity (12,21,23-26)
The correlation between DPYD*9A (c.85T>C) genotype and DPD deficiency clinical phenotype is controversial (27,28). Reference laboratories either did not perform DPYD*9A genotyping or have stopped DPYD*9A genotyping and limited genotyping to high-risk variants (DPYD*2A, DPYD*13 and DPYD*9B) only. DPYD*9A (c.85T>C) variant was the most common variant diagnosed in our cohort and a genotype-clinical phenotype correlation was noticeable. Thus, here we explored DPYD*9A genotype and clinical phenotype correlation in patients with GI malignancies treated with fluoropyrimidines.
Methods
Patient population
This is a retrospective study conducted at the University of South Alabama Mitchell Cancer Institute in Mobile, Alabama, USA. Cohort was identified through searching our cancer center tumor registry for patients genotyped for DPYD variants between 2011 and 2017. The University of South Alabama Institutional Review Board (IRB) approved this study and the IRB-approved database provided a waiver of the requirement for informed consent and allowed for publication of de-identified data (IRB #836682-3).
DPYD Genotyping
Germline DNA was obtained from peripheral blood specimen and genotyped for DPYD variants in ARUP laboratories (Salt lake city, UT, USA) or LabCorp laboratories (Burlington, NC, USA) depending on patients’ specific health insurance. Between 2011 and 2014, ARUP laboratories provided results for 5 variants [IVS14+1G>A (DPYD*2A), DPYD c.1679T>G (DPYD*13A), DPYD c.2846A>T (DPYD*9B), DPYD c.85T>C (DPYD*9A) and DPYD c.1590T>C]. Between 2015 and 2017, ARUP laboratories provided results for only 3 variants [IVS14+1G>A (DPYD*2A), DPYD c.1679T>G (DPYD*13A) and DPYD c.2846A>T (DPYD*9B)]. LabCorp Laboratories provided results for IVS14+1G>A (DPYD*2A) only. When a mutation was identified, heterozygous or homozygous status was included in the final result report.
Toxicity grading and statistical analysis
Demographic and clinical data were extracted from the patients’ charts. Toxicity was graded according to the National Cancer Institute Common Terminology Criteria for Adverse Events (version 3.0) (29). Association between dichotomous fluoropyrimidine-related toxicities and DPYD variants status was performed using Fisher’s exact test. Analyses with P values ≤0.05 were considered significant. Tests were performed using GraphPad software QuickCalcs (GraphPad software 2016, San Diego, California, USA).
Results
Patients characteristics
Between 2011 and 2017, a total of 67 patients with GI malignancies were genotyped for DPYD variants. The baseline characteristics of the patients are summarized in Table 1. Median age is 60 years. Males represented 51% of the patients while females represented 49%. In our cohort, 76% were Caucasian, 22% were African Americans and 2% were Hispanics. Colon adenocarcinoma represented the most common malignancy in our cohort. Other patients had anal squamous cell carcinoma (SCC), cholangiocarcinoma, esophageal adenocarcinoma, gastric adenocarcinoma, neuroendocrine tumor of the small bowel (jejunum), pancreatic adenocarcinoma and rectal adenocarcinoma. A Fluorouracil-based chemotherapy regimen was administered in 34 (51%) patients while 33 (49%) patients received a capecitabine-based chemotherapy regimen.

Full table
DPYD Genotyping
In 24 patients (36%), the treating oncologist considered DPYD genotyping prior to the initiation of treatment with fluoropyrimidines. Such decision to perform upfront genotyping was mainly done due to concerns about patients’ fitness to handle potential toxicities due to their age or the presence of significant comorbidities. The other, 43 patients (64%), were genotyped for DPYD after they have experienced grade 3 or greater toxicities.
All patients (N=67) were genotyped for IVS14+1G>A (DPYD*2A) variant. Genotyping for DPYD c.1679T>G (DPYD*13A) and DPYD c.2846A>T (DPYD*9B) variants was performed in 55 patients (82%) only. Genotyping for DPYD c.85T>C (DPYD*9A) and DPYD c.1590T>C variants was performed in 28 patients (42%) only. The strategy of genotyping by the treating oncologist and the DPYD variants genotyped are summarized in Table 2.

Full table
DPYD genotyping analysis
The genotyping analysis of the patients included in our cohort is summarized in Table 2. DPYD variants were identified in 17 out of 67 patients (25%). One patient was homozygous for DPYD*9A (c.85T>C) variant and one patient was double heterozygous for DPYD*9A (c.85T>C) and DPYD*9B variants. The remaining of the patients were heterozygous. Among the patients with identified DPYD variants (N=17), 13 (76%) patients had DPYD c.85T>C (DPYD*9A) variant, 3 (18%) patients had DPYD IVS14+1G>A (DPYD*2A) variant and 2 (12%) patient had DPYD c.2846A>T (DPYD*9B) variant. Neither DPYD*13A nor DPYD c.1590T>C variants were identified.
Only 28 patients were genotyped for DPYD*9A (c.85T>C) variant since LabCorp laboratories did not perform DPYD*9A genotyping and since ARUP laboratories have stopped DPYD*9A genotyping and limited genotyping to high-risk variants (DPYD*2A, DPYD*13 and DPYD*9B) only. Among patients screened for DPYD*9A (c.85T>C) variant (N=28), 13 patients (46%) had DPYD*9A (c.85T>C) variant and 1 (4%) patient had DPYD IVS14+1G>A (DPYD*2A) variant. None of the other variants (DPYD*9B, DPYD*13A and DPYD c.1590T>C) were identified.
Adverse events
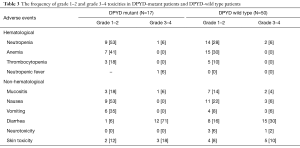
The frequency of grade 1–2 and grade 3–4 toxicities in DPYD-mutant patients and DPYD-wild type patients is summarized in Table 3. None of the patients have died as a consequence of fluoropyrimidines-induced toxicities. The most common experienced grade 3–4 toxicity in both DPYD-mutant patients and DPYD-wild type patients was diarrhea.
Full table
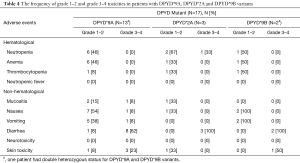
The frequency of grade 1–2 and grade 3–4 toxicities in patients with DPYD*9A, DPYD*2A and DPYD*9B variants is summarized in Table 4. In our cohort, 8 patients out of 13 patients (62%) with DPYD*9A (c.85T>C) variant developed grade 3–4 diarrhea. The five patients who did not develop grade 3–4 diarrhea received dose reduced chemotherapy as they were genotyped by the treating oncologist prior to initiation of treatment. All patients with DPYD*2A (N=3) and DPYD*9B (N=2) received full dose chemotherapy and experienced grade 3–4 diarrhea. Of note, 3 patients with DPYD*9A variant developed grade 3–4 skin toxicity. The skin toxicity in 2 of those 3 patients was manifested as balanitis.
Full table
Statistical analysis
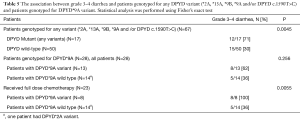
In all patients, grade 3–4 diarrhea was associated with DPYD mutant status (any variant) (P=0.0045). In patients genotyped for DPYD*9A (c.85T>C) variant (N=28), patients with DPYD*9A (c.85T>C) variant tend to experience more grade 3–4 diarrhea (62%) compared to patients with DPYD*9A (c.85T>C) wild-type (36%). However, the association between DPYD*9A (c.85T>C) status and grade 3–4 diarrhea did not reach statistical significance (P=0.256) likely due to small sample size. In patients genotyped for DPYD*9A and received full dose fluoropyrimidines (N=23), grade 3–4 diarrhea was associated with DPYD*9A (c.85T>C) variant (P=0.0055). The association between grade 3–4 diarrhea and patients genotyped for any DPYD variant (*2A, *13A, *9B, *9A and /or DPYD c.1590T>C) and patients genotyped for DPYD*9A (c.85T>C) variant is summarized in Table 5.
Full table
Discussion
The correlation between DPYD*9A (c.85T>C) genotype and impaired DPD activity has been demonstrated in laboratory and clinical studies. In Escherichia coli, DPYD*9A (c.85T>C) mutation lead to a mutant DPD protein (C29R) leading to significant decrease in enzymatic activity (30). In patients with DPYD*9A (c.85T>C) variant, DPD activity has been reported to be decreased (27,31,32). In one study of patients with GI malignancies, DPYD*9A (c.85T>C) variant was associated with fluoropyrimidines-associated toxicity. Patients experienced diarrhea (P<0.05) and hand foot syndrome (HFS) (P<0.05) (27).
Understandably so, the correlation between DPYD*9A (c.85T>C) genotype and impaired DPD activity continued to be controversial as other clinical studies reported no correlation between DPYD*9A (c.85T>C) genotype and DPD deficiency clinical phenotype (33-35). Moreover, additional studies suggested that DPYD*9A (c.85T>C) may serve as a protective allele against fluoropyrimidines-associated toxicity (28). Based on the current limited knowledge, the 2017 updated Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for DPD genotype and fluoropyrimidine dosing, it was stated that the DPYD*9A (c.85T>C), among other variants, doesn’t affect DPD activity in a clinically relevant manner (36).
In our cohort, a correlation between the DPYD*9A (c.85T>C) variant genotype and DPD deficiency clinical phenotype was noticeable. All patients (N=8) who received full dose fluoropyrimidines experienced grade 3–4 diarrhea. Among those patients, three patients, in addition to diarrhea, developed skin toxicity manifested as balanitis in two patients and HFS in one patient. None of the patients with DPYD*9A (c.85T>C) variant developed grade 3–4 hematological toxicities.
In our cohort of patients with DPYD*9A (c.85T>C) variant (N=13), only one patient was found to be homozygous for DPYD*9A. This patient experienced grade 3 diarrhea on day 3 of cycle #1 adjuvant XELOX given for stage III colon adenocarcinoma. Capecitabine was stopped immediately after the experienced toxicity. Due to toxicity, the planned adjuvant chemotherapy (XELOX) was discontinued and the patient was started on surveillance. Unfortunately, the patient developed disease recurrence 2 years later. Since then alternate regimens that omit fluoropyrimidines (IROX and bevacizumab) and trifluridine and tipiracil have been used.
One patient was found to be double heterozygous for DPYD (DPYD*9A (c.85T>C) and DPYD*9B). This patient experienced grade 3 balanitis toward the end of neoadjuvant concurrent chemoradiation with capecitabine given for stage II rectal adenocarcinoma. DPYD genotyping was not considered at that time. After surgical resection, the patient was started on adjuvant chemotherapy with XELOX. Shortly after cycle #2, he experienced grade 3 diarrhea. Genotyping for DPYD was considered then and revealed double heterozygous status. The treating oncologist continued the planned adjuvant chemotherapy but he reduced the dose of capecitabine by 50%. The patient completed the rest of the planned adjuvant chemotherapy and it was well tolerated. Patient continues to do well without evidence of disease recurrence.
The remaining eleven patients were found to be heterozygous for DPYD variants. Five patients were diagnosed to have heterozygous DPYD*9A (c.85T>C) variant prior to initiation of therapy. In those patients fluoropyrimidines-based regimens were administered at a reduced dose followed by dose adjustment at the discretion of the treating oncologist discretion. Treatment was well tolerated. Six patients were diagnosed to have heterozygous DPYD*9A (c.85T>C) variant after initiation of full dose fluoropyrimidines-based regimens and after they experienced grade 3–4 toxicity. Subsequently, fluoropyrimidines-based regimens were administered at a reduced dose followed by dose adjustment at the discretion of the treating oncologist discretion. Treatment was well tolerated.
In addition to the noticeable correlation between DPYD*9A (c.85T>C) variant genotype and clinical phenotype of DPD deficiency, our study represents one other study that shows the association between diarrhea and DPYD*9A (c.85T>C) variant. Joerger et al., reported that grade 1–4 diarrhea was associated with DPYD*9A (c.85T>C) in patients with gastroesophageal cancer (p=0.0023). In the same study grade 1–4 HFS was associated with DPYD*9A (c.85T>C) variant in patients with colorectal cancer (P=0.0033) (27).
The current genotyping strategy adopted by ARUP laboratories (that limits genotyping to high risk variants only) and LabCorp (that limits genotyping to DPYD*2A variant only) has several limitations. High-risk variants (DPYD *2A, *13 and *9B) were identified in only 30% of patients who developed grade 3–4 toxicities after exposure to fluoropyrimidines (21). The Clinical Pharmacogenetics Implementation Consortium concluded that 23–38% of severe fluoropyrimidines-associated toxicity could be attributed to DPYD variants (clinical sensitivity approximately 31%) (37). In a prospective study, DPYD*2A variant was identified in only 5% of patients experiencing grade 3–4 fluoropyrimidines-associated toxicity. Moreover, less than 50% of patients with DPYD*2A variant developed grade 3–4 toxicity (the positive predictive value was 46%) (26). In another study, only 6% of patients experiencing grade 3–4 toxicity had a high-risk mutant DPYD variant but the positive predictive value was >99% (38). Moreover, it is important to recognize that DPD deficiency has been identified in patients with wild-type DPYD alleles probably due to epigenetic mechanisms (39,40).
The limitations observed by the current adopted genotyping strategy, that limit screening to DPYD*2A variant only or high-risk variants (DPYD *2A, *13 and *9B), are important to recognize. The patients treated with fluoropyrimidines and their treating oncologist would certainly benefit from adopting a better screening approach for DPD deficiency. The controversy about the correlation between DPYD*9A (c.85T>C) genotype and DPD deficiency clinical phenotype is understandable. However, with the available data that show DPYD*9A (c.85T>C) genotype-clinical phenotype correlation, the decision of not including DPYD*9A (c.85T>C) variant in the screening panel maybe premature at this point. Our study showed that DPYD*9A (c.85T>C) variant genotyping has helped medical oncologists in the clinic to identify patients with an underlying genetic alteration that could predispose them to experience grade 3–4 fluoropyrimidines-associated toxicity. If our cohort was only genotyped for DPYD*2A or high-risk variants only, many patients would have been undiagnosed and their outcomes could have been different. By including DPYD*9A (c.85T>C) variant to the screening panel by reference laboratories, the oncology community would have more data about its clinical significance.
Our study has several limitations. This study is a retrospective study and there are inherent limitations and selection bias associated with a retrospective analysis of this sort, particularly regarding the oncologist discretion to conduct upfront screening in particular subjects. In 24 patients (36%), the treating oncologist considered DPYD genotyping prior to the initiation of treatment with fluoropyrimidines mainly due to concerns about patients’ fitness to handle potential toxicities due to their age or the presence of significant comorbidities. Five patients with DPYD*9A variants were genotyped prior to initiation of treatment.
This study represents a single institution experience and the sample size is relatively small. Moreover, there is significant heterogeneity in the patient population, including inclusion of multiple different tumor types and a variety of fluoropyrimidine-based regimens (many of which were, presumably, chemotherapy combinations that may have differentially contributed to the observed toxicity). Despite the correlative findings that we were able to generate, studies of larger cohorts and ideally conducted prospectively will likely provide more solid data about such genotype and clinical phenotype correlation. Overall, our study should be considered hypothesis-generating rather than definitive due to its limitations.
Conclusions
DPD enzyme deficiency is a pharmacogenetic syndrome associated with dose-limiting toxicity to fluoropyrimidines. In our cohort, DPYD*9A (c.85T>C) variant was the most common diagnosed variant. The correlation between DPYD*9A genotype and DPD deficiency clinical phenotype was noticeable in patients who received full dose fluoropyrimidines as they all experienced grade 3–4 toxicities (diarrhea). Oncologists should consider DPYD genotyping for patients experiencing grade 3–4 diarrhea after exposure to fluoropyrimidines. The prevalence of partial DPD deficiency in the general population is approximately 3–5%. Genotyping for high-risk DPYD variants (*2A, 13A and 9B) only has several limitations and is suboptimal as it leaves most patients with DPD deficiency undiagnosed. A more comprehensive approach would include testing for additional DPYD variants [including but not limited to DPYD*9A (c.85T>C) variant] or ideally screening the entire coding region and potentially additional regions responsible for regulating DPYD gene expression and translation. Despite the correlative findings that we were able to generate, studies of larger cohorts and ideally conducted prospectively will likely provide more data about DPYD*9A genotype and clinical phenotype correlation.
Acknowledgements
Authors thankfully acknowledge the USA-MCI for the consent to access the patient’s genotyping data and IRB, to allow the publication of de-identified data.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The University of South Alabama Institutional Review Board (IRB) (No. 836682-3) approved this study and the IRB-approved database provided a waiver of the requirement for informed consent and allowed for publication of de-identified data.
References
- Miwa M, Ura M, Nishida M, et al. Design of a novel oral fluoropyrimidine carbamate, capecitabine, which generates 5-fluorouracil selectively in tumours by enzymes concentrated in human liver and cancer tissue. Eur J Cancer 1998;34:1274-81. [Crossref] [PubMed]
- Huang WY, Ho CL, Lee CC, et al. Oral tegafur-uracil as metronomic therapy following intravenous FOLFOX for stage III colon cancer. PLoS One 2017;12:e0174280. [Crossref] [PubMed]
- Ershler WB. Capecitabine monotherapy: safe and effective treatment for metastatic breast cancer. Oncologist 2006;11:325-35. [Crossref] [PubMed]
- Vermorken JB, Remenar E, van Herpen C, et al. Cisplatin, fluorouracil, and docetaxel in unresectable head and neck cancer. N Engl J Med 2007;357:1695-704. [Crossref] [PubMed]
- Thorn CF, Marsh S, Carrillo MW, et al. PharmGKB summary: fluoropyrimidine pathways. Pharmacogenet Genomics 2011;21:237-42. [Crossref] [PubMed]
- Hoff PM, Ansari R, Batist G, et al. Comparison of oral capecitabine versus intravenous fluorouracil plus leucovorin as first-line treatment in 605 patients with metastatic colorectal cancer: results of a randomized phase III study. J Clin Oncol 2001;19:2282-92. [Crossref] [PubMed]
- Koopman M, Antonini NF, Douma J, et al. Sequential versus combination chemotherapy with capecitabine, irinotecan, and oxaliplatin in advanced colorectal cancer (CAIRO): a phase III randomised controlled trial. Lancet 2007;370:135-42. [Crossref] [PubMed]
- Van Cutsem E, Twelves C, Cassidy J, et al. Oral capecitabine compared with intravenous fluorouracil plus leucovorin in patients with metastatic colorectal cancer: results of a large phase III study. J Clin Oncol 2001;19:4097-106. [Crossref] [PubMed]
- Tuchman M, Stoeckeler JS, Kiang DT, et al. Familial pyrimidinemia and pyrimidinuria associated with severe fluorouracil toxicity. N Engl J Med 1985;313:245-9. [Crossref] [PubMed]
- Heggie GD, Sommadossi JP, Cross DS, et al. Clinical pharmacokinetics of 5-fluorouracil and its metabolites in plasma, urine, and bile. Cancer Res 1987;47:2203-6. [PubMed]
- Saif MW, Syrigos K, Mehra R, et al. Dihydropyrimidine Dehydrogenase Deficiency (Dpd) in Gi Malignancies: Experience of 4-Years. Pak J Med Sci 2007;23:832-9. [PubMed]
- Lee AM, Shi Q, Pavey E, et al. DPYD variants as predictors of 5-fluorouracil toxicity in adjuvant colon cancer treatment (NCCTG N0147). J Natl Cancer Inst 2014;106:dju298. [Crossref] [PubMed]
- van Kuilenburg AB, Haasjes J, Richel DJ, et al. Clinical implications of dihydropyrimidine dehydrogenase (DPD) deficiency in patients with severe 5-fluorouracil-associated toxicity: identification of new mutations in the DPD gene. Clin Cancer Res 2000;6:4705-12. [PubMed]
- Mattison LK, Fourie J, Desmond RA, et al. Increased prevalence of dihydropyrimidine dehydrogenase deficiency in African-Americans compared with Caucasians. Clin Cancer Res 2006;12:5491-5. [Crossref] [PubMed]
- Etienne MC, Milano G, Renee N, et al. Population study of dihydropyrimidine dehydrogenase in cancer patients. Bull Cancer 1995;82:705-10. [PubMed]
- Lu Z, Zhang R, Diasio RB. Dihydropyrimidine dehydrogenase activity in human peripheral blood mononuclear cells and liver: population characteristics, newly identified deficient patients, and clinical implication in 5-fluorouracil chemotherapy. Cancer Res 1993;53:5433-8. [PubMed]
- van Kuilenburg AB, De Abreu RA, van Gennip AH. Pharmacogenetic and clinical aspects of dihydropyrimidine dehydrogenase deficiency. Ann Clin Biochem 2003;40:41-5. [Crossref] [PubMed]
- Lu Z, Zhang R, Carpenter JT, et al. Decreased dihydropyrimidine dehydrogenase activity in a population of patients with breast cancer: implication for 5-fluorouracil-based chemotherapy. Clin Cancer Res 1998;4:325-9. [PubMed]
- Raida M, Schwabe W, Hausler P, et al. Prevalence of a common point mutation in the dihydropyrimidine dehydrogenase (DPD) gene within the 5'-splice donor site of intron 14 in patients with severe 5-fluorouracil (5-FU)- related toxicity compared with controls. Clin Cancer Res 2001;7:2832-9. [PubMed]
- Yamaguchi K, Arai Y, Kanda Y, et al. Germline mutation of dihydropyrimidine dehydrogenese gene among a Japanese population in relation to toxicity to 5-Fluorouracil. Jpn J Cancer Res 2001;92:337-42. [Crossref] [PubMed]
- Morel A, Boisdron-Celle M, Fey L, et al. Clinical relevance of different dihydropyrimidine dehydrogenase gene single nucleotide polymorphisms on 5-fluorouracil tolerance. Mol Cancer Ther 2006;5:2895-904. [Crossref] [PubMed]
- Offer SM, Fossum CC, Wegner NJ, et al. Comparative functional analysis of DPYD variants of potential clinical relevance to dihydropyrimidine dehydrogenase activity. Cancer Res 2014;74:2545-54. [Crossref] [PubMed]
- Offer SM, Lee AM, Mattison LK, et al. A DPYD variant (Y186C) in individuals of african ancestry is associated with reduced DPD enzyme activity. Clin Pharmacol Ther 2013;94:158-66. [Crossref] [PubMed]
- Johnson MR, Wang K, Diasio RB. Profound dihydropyrimidine dehydrogenase deficiency resulting from a novel compound heterozygote genotype. Clin Cancer Res 2002;8:768-74. [PubMed]
- Boisdron-Celle M, Remaud G, Traore S, et al. 5-Fluorouracil-related severe toxicity: a comparison of different methods for the pretherapeutic detection of dihydropyrimidine dehydrogenase deficiency. Cancer Lett 2007;249:271-82. [Crossref] [PubMed]
- Schwab M, Zanger UM, Marx C, et al. Role of genetic and nongenetic factors for fluorouracil treatment-related severe toxicity: a prospective clinical trial by the German 5-FU Toxicity Study Group. J Clin Oncol 2008;26:2131-8. [Crossref] [PubMed]
- Joerger M, Huitema AD, Boot H, et al. Germline TYMS genotype is highly predictive in patients with metastatic gastrointestinal malignancies receiving capecitabine-based chemotherapy. Cancer Chemother Pharmacol 2015;75:763-72. [Crossref] [PubMed]
- Kleibl Z, Fidlerova J, Kleiblova P, et al. Influence of dihydropyrimidine dehydrogenase gene (DPYD) coding sequence variants on the development of fluoropyrimidine-related toxicity in patients with high-grade toxicity and patients with excellent tolerance of fluoropyrimidine-based chemotherapy. Neoplasma 2009;56:303-16. [Crossref] [PubMed]
- National Cancer Institute: Common Termi- nology Criteria for Adverse Events v3.0. Available online: http://ctep. cancer.gov/protocolDevelopment/electronic_applications/ docs/ctcaev3.pdf
- Vreken P, van Kuilenburg AB, Meinsma R, et al. Dihydropyrimidine dehydrogenase deficiency: a novel mutation and expression of missense mutations in E. coli. J Inherit Metab Dis 1998;21:276-9. [Crossref] [PubMed]
- Vreken P, Van Kuilenburg AB, Meinsma R, et al. Dihydropyrimidine dehydrogenase (DPD) deficiency: identification and expression of missense mutations C29R, R886H and R235W. Hum Genet 1997;101:333-8. [Crossref] [PubMed]
- Van Kuilenburg AB, Vreken P, Riva D, et al. Clinical and biochemical abnormalities in a patient with dihydropyrimidine dehydrogenase deficiency due to homozygosity for the C29R mutation. J Inherit Metab Dis 1999;22:191-2. [Crossref] [PubMed]
- Amstutz U, Farese S, Aebi S, et al. Dihydropyrimidine dehydrogenase gene variation and severe 5-fluorouracil toxicity: a haplotype assessment. Pharmacogenomics 2009;10:931-44. [Crossref] [PubMed]
- Gross E, Busse B, Riemenschneider M, et al. Strong association of a common dihydropyrimidine dehydrogenase gene polymorphism with fluoropyrimidine-related toxicity in cancer patients. PLoS One 2008;3:e4003. [Crossref] [PubMed]
- Seck K, Riemer S, Kates R, et al. Analysis of the DPYD gene implicated in 5-fluorouracil catabolism in a cohort of Caucasian individuals. Clin Cancer Res 2005;11:5886-92. [Crossref] [PubMed]
- Amstutz U, Henricks LM, Offer SM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing: 2017 update. Clin Pharmacol Ther 2018;103:210-6. [Crossref] [PubMed]
- Caudle KE, Thorn CF, Klein TE, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing. Clin Pharmacol Ther 2013;94:640-5. [Crossref] [PubMed]
- Loganayagam A, Arenas Hernandez M, Corrigan A, et al. Pharmacogenetic variants in the DPYD, TYMS, CDA and MTHFR genes are clinically significant predictors of fluoropyrimidine toxicity. Br J Cancer 2013;108:2505-15. [Crossref] [PubMed]
- Ezzeldin HH, Lee AM, Mattison LK, et al. Methylation of the DPYD promoter: an alternative mechanism for dihydropyrimidine dehydrogenase deficiency in cancer patients. Clin Cancer Res 2005;11:8699-705. [Crossref] [PubMed]
- Amstutz U, Offer SM, Sistonen J, et al. Polymorphisms in MIR27A associated with early-onset toxicity in fluoropyrimidine-based chemotherapy. Clin Cancer Res 2015;21:2038-44. [Crossref] [PubMed]


