Detecting promoter methylation pattern of apoptotic genes Apaf1 and Caspase8 in gastric carcinoma patients undergoing chemotherapy
Introduction
Gastric cancer is the fourth most common cancer in which is the second leading cause of death-related cancers (1,2). Its incidence varies among different regions and countries. Each year, there are approximately 934,000 new cases of gastric cancer in the world, 56% of which occur in East Asia. Among these new cases, 41% come from China and 11% from Japan (3,4). There are two main types of gastric adenocarcinoma: intestinal and diffuse. The accepted paradigm for the pathogenesis of the intestinal-type is a multi-step progress from chronic gastritis to gastric atrophy to intestinal metaplasia to dysplasia. The pathogenesis of diffuse-type gastric cancer is not yet completely understood (5).
Both environmental alternations (6) including Helicobacter pylori (H. pylori) infection (7), Epstein-Barr virus (8), diet (5) and Genetic alterations, such as p53, KRAS, PIK3CA, ARID1A, MLL3 and MLL mutations, as well as PIK3CA, C-MET, ERBB4, and CD44 amplifications, are repeatedly found in gastric cancer, that suggest key tumorigenic events and their critical role in gastric tumor genesis (9,10). Epigenetic alterations also involved in progression of gastric cancer, including DNA methylation, post-translational modifications of histones, noncoding RNAs, and nucleosome positioning (11,12). DNA methylation which is the most common epigenetic alternation occurs at the C5 position of cytosine (5mC), mostly within CpG dinucleotide, with the DNMT enzymes using a protected mechanism, which provides a stable gene silencing mechanism and it has an important role in regulation of gene expression (13-15). DNA methylation can grant a selective growth advantage to cells when it occurs in the promoter regions of genes involved in growth regulation and DNA damage responses, that results in the development of cancer (16).
Gastric cancer’s formation and progression are processes which are continuous and multiple-step (17,18). The best-known type of programmed cell death is apoptosis which plays important roles in growth and homeostasis as well as pathogenesis of many diseases (19,20). The apoptotic cascade can be triggered through two major pathways extracellular signals, such as members of the tumor necrosis factor (TNF) family can activate the receptor-mediated extrinsic pathway. Alternatively, stress signals such as DNA damage, hypoxia, and loss of survival signals may trigger the mitochondrial intrinsic pathway (21).
Various cell death receptors including TNFR1 and CD95L can trigger the caspase-8 dependent extrinsic apoptotic pathway (22,23). One of the best-defined apoptotic pathways is mediated by the death receptor CD95 (APO-1/Fas). Triggering of CD95 by its natural ligand CD95L or agonistic antibodies induces the formation of a death-inducing signaling complex (DISC) consisting of the adaptor protein Fas-associated death domain protein [FADD (MORT-1)] and procaspase-8 [FADD-like IL-1 b–converting enzyme (FLICE, Mch5)] (24,25). Two hemophilic protein interaction domains mediate the DISC formation: FADD which contains a COOH-terminal death domain (DD), and couples to the DD of the intracellular part of CD95. FADD, in addition, contains an NH2-terminal so-called death effector domain (DED), which binds to one of the DEDs of caspase-8. Further downstream, caspase-8 triggers the proteolysis activation of other caspases and cleavage of cellular substrates (21,26). Apaf-1 gene participates in the pathway of mitochondria-mediated apoptosis. UV and ionizing radiation, hypoxia, cytochrome c is released from the mitochondria when DNA is damaged by chemotherapeutic agents, oncogenic stimuli, binds to Apaf-1 in the cytosol, and in association with dATP/ATP, facilitates a conformational change of Apaf-1 to expose its CARD domain (27). Afterwards, Apaf-1 oligomerizes through the unconcealed CARD domain and catalyzes auto activation of caspase-9, leading to the serial activation of downstream effector caspases such as caspase-3, caspase-6, and caspase-7, which results in apoptotic cell death (28).
Given the importance of Apaf1 and Casp8 genes in the process of apoptosis in patients receiving chemotherapy, the aim of this study was to investigate the relationship between patterns of apoptotic gene promoter methylation in gastric carcinoma in patients undergoing chemotherapy. In addition, Comparison of the promoter methylation in the blood and tissue samples in gastric cancer was performed and the impact of epigenetic in carcinogenesis was examined. Also in this study the relationship between methylation patterns of these genes with clinicopathological characteristics of patients was investigated.
Methods
Study population
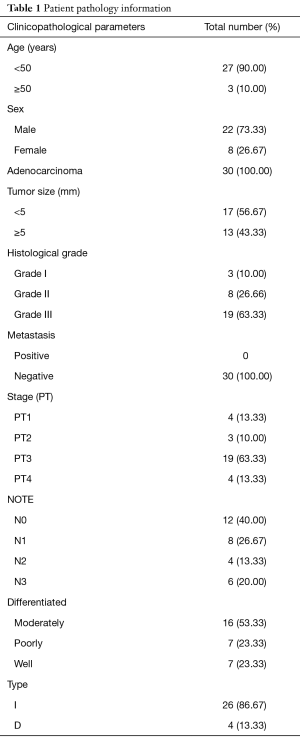
Thirty samples of patient’s blood and tissue that were diagnosed through clinical experiments of gastric carcinoma with average age of 61.8 years old were provided from Imam Khomeini (mercy upon him) hospital. 30 tissue samples of control individuals without any record of gastric carcinoma or related clinical symptoms and no kinship relations with patients were selected from Imam Hussein (PBH) hospital (pathological data were indicated in Table 1). Tissue samples were paraffinized after surgery which maintainable in laboratory temperature, blood samples were provided in the tube containing EDTA and maintained for a longtime in −20 °C. This study was conducted at the Biological Research Center of Azad Islamic University of Zanjan and approved by the faculty of medical sciences Ethics Committee. Informed consent was taken from all the patients before entering the study and all the obtained information’s from each participant was completely confidential.
Full table
Analysis of CASP8 and Apaf1 promoter methylation status
Genomic DNA was extracted from 25–30 ng of tissue using a ZS Genomic DNA™ Tissue Mini Prep Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The DNA concentration was determined by spectrophotometry, and its integrity was checked by 1% gel electrophoresis. Bisulfite treatment was performed using an EZ DNA Methylation Gold Kit™ (Qiagen, Hilden, Germany) according to the manufacturer’s instructions/protocol. The methylation status of the promoters was detected by methylation-specific polymerase chain reaction (MSP). The methylated and unmethylated DNA sequence primers are listed in Table 2. PCR was performed in a total volume of 20 µL, containing 10 µL (1×). Master Mix (PCR buffer, dNTP, MgCl2, Taq DNA polymerase), 6 µL DNase Free Water, 1 µL (0.5 µM) Forward primer, 1 µL (0.5 µM). Reverse primer and 2 µL (100 ng) of converted DNA. PCR cycling conditions were: initial denaturation at 95 °C for 5 min, followed by 35 cycles of 95 °C denaturation for 45 s, annealing for 45 s (primer specific temperatures are listed in Table 2), 72 °C extensions for 45 s, and final extension at 72 °C for 5 min. The PCR products were separated by 2.5% gel electrophoresis. If both methylated and non-methylated bands appeared in the gel represents the hemi-methylation status, if only methylated or non-methylated bands appeared, fully methylated and non-methylated situation was confirmed respectively.

Full table
Statistical analysis
Statistical analysis was carried out with the SPSS20 Statistics software. Quantitative data are presented as mean and standard deviation (SD). For testing statistical hypothesis about the independence of two variables, the chi-square test or the Fisher exact test was used. A Spearman coefficient was calculated to determine correlation. The significance level of <0.05 was selected.
Results
Characterization of clinical specimens
According to the expert diagnosis of pathological analysis, all of the patients confirmed with gastric cancer and in the process of metastasis. As shown in Table 1, the disease is about 73.33% men and 26.67% of women and 90% are younger than 50 years and most of intestinal type is involved. The majority of patients had tumor size less than 5 mm as well as 63.33% of patients with histological grade III and 63.33% in stage III disease, and also the majority of patients in the moderately and 40% of them are in stage Note0.
Methylation analysis
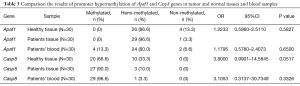
The methylation status of the promoter region of Apaf1 and Casp8 genes was analyzed in 60 FFPE samples (30 cancer cases and 30 normal cases) and 30 blood cancer samples. Thirty cases were analyzed for blood and tissue samples from the same patient simultaneously. Methylation frequencies of Apaf1 and Casp8 genes in tumor and normal tissues and blood samples have been shown in Table 3. According to the table, tissue samples analysis that Apaf1 and casp8 genes promoter in normal FFPE samples were methylated (m+/u–) in 0% and 66.6%, respectively; hemimethylated (m+/u+) in 86.6% and 33.3%, and non-methylated in 13.3% and 0%, respectively; in comparison to patient FFPE sample were methylated 0% and 90%, hemimethylated in 96.6% and 10%, and non-methylated in 3.3% and 0%, our data confirmed significant relationship between promoter hyper methylation of casp8 gene and gastric cancer (P<0.05). It can be concluded that no significant association between promoter methylation of two genes at the same time (P>0.05).
Full table
Also according to analyze the correlation between patient’s tissue and blood samples of Apaf1 and Casp8 genes, methylation frequency showed (0%, 13.3% and 90%, 96.6%), hemimethylation (96.6%, 80% and 10%, 3.3%), unmethylation (3.3%, 6.6% and 0%, 0%) respectively. There is no significant relationship between methylation frequency of Apaf1 and Casp8 genes in tissue and blood samples of patients (P=0.6 and 0.3). So evaluating the status of methylation in the blood offered as a non-invasive approach.
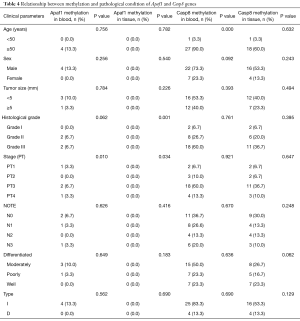
Clinicopathological parameters of gastric cancer were compared with the frequency of Apaf1 and Casp8 genes promoter methylation in Table 4. Our results from this analysis indicated there is a significant relationship between Apaf1 gene methylation in blood with stage of cancer (P<0.05) and methylation of this gene in tissue with stage (P<0.05) and grade (P<0.01). In addition correlation between promoter hypermethylation of Casp8 gene in blood and age of patients, significant association has been observed (P<0.001), but no significant relationship was seen in other pathological factors.
Full table
Discussion
DNA methylation is the most extensively and widely studied epigenetic modification in which a methyl group is added to the fifth carbon position of cytosine residue in a CpG dinucleotide. Clusters of CpG dinucleotides in CG rich regions of the genome called “CpG islands (CGI)” frequently occurs in the 5'-flanking promoter areas of genes (29). Generally, increased methylation in the promoter region of genes leads to reduced gene expression, whereas methylation in the transcribed region has a variable effect on gene expression (30,31). So, any perceptive understanding of abnormal methylation and subsequent gene silencing, such as methylation inducing factors, which is essential for cancer prediction, prevention, treatment and prognosis evaluation (32).
The empirical evidence showed that the percentage of Apaf1 gene methylation in normal tissue and the patient and also the percentage of methylation of this gene in tissue and blood with P>0.05, there was not a significant relationship. Li and colleagues investigated the role of DNA methylation in 2003 to prevent Apaf1 protein expression in human leukemia addressed in this study promoter methylation of Apaf1 in four cases were examined and it was found that P<0.05 was significant. There was evidence that showed Apaf1 gene methylation detection and treatment by demethylation, regulates Apaf1 positive expression in both protein and mRNA expression (33). In another study, Wang and associates in 2007 evaluated Apaf1 gene expression in gastric cancer of 35 samples of cancer tissue and adjacent normal tissue samples, methylation detection was performed by MSP method. Promoter methylation rate was detected 49% in gastric cancer tissue. Methylation significantly decreased expression in 16 of 18 cancerous tissue samples (P=0.000001) (34). In another study, Huang and colleagues in 2004 studied Apaf1 gene promoter methylation in squamous cell carcinoma of the larynx and in all 11 samples showed that the methylation of promoter regions decreased mRNA gene expression (35). But in the current research, no significant association between gene methylation of 30 patient tissue samples and 30 normal tissue samples with methylation specific PCR method does exist.
In this study we did found that the percentage of Casp8 gene methylation in normal and pathological tissue as well as tissue between the percentage of methylation of this gene in the blood of patients with P<0.05 there was a significant relationship. Rita and colleagues in 2013 examined methylation CpG islands of Casp8 gene in tumor tissues and adjacent non-cancerous stomach tissues. In this study, gene methylation of Casp8 in 69 patients with gastric cancer using methylation-specific PCR was performed, the frequency of methylation in cancer and non-cancerous adjacent was (5.8%), therefore correlation between the methylation of this gene and gastric cancer does not exist (36). Skiriute and colleagues in another study in 2012 examined promoter methylation status of Casp8 gene for 76 patients with glioblastoma using MSP, they found 56.8% methylated (37). As well as Kordi Tamandani and colleagues in a study in 2009 assess the methylation of CpG islands of Casp8 gene using methylation specific PCR method in 80 patients with cervical cancer. The rate of methylation was 1.2% and 1.80% respectively with P>0.05 found a significant association between gene methylation of CpG sites and cervical cancer (38). In our study by MSP on tissue samples from 30 patients and 30 normal tissues showed that significant correlation between the percentage of methylation of Casp8 gene and gastric cancer.
Pathological results of this analysis showed a significant correlation between the percentage of Apaf1 gene methylation in blood and tumor stage (P<0.05). Communication between methylation in the context of the stage (P<0.05) and grade (P<0.01), there was a significant relationship, But relationship between other pathological characteristics such as age, sex, tumor size, grade has not been seen. There was also a significant association between Casp8 gene methylation in blood and age (P<0.001), but there was not a significant relationship with other pathological information such as gender, tumor size, Stage, grade. Kupcinskaite-Noreikiene and colleagues in 2013 examined CpG islands methylation of Casp8 gene in tumor tissues and adjacent non-cancerous tissues in the stomach. In this study, gene methylation in 69 patients with gastric cancer using methylation-specific PCR was performed, results confirmed that the frequency of methylation in cancer and non-cancerous adjacent was 8.5%, therefore no correlation was detected between methylation of the gene and gastric cancer, no link between the characteristics of the patients and methylation of this gene was investigated. And it was found that a significant correlation between the pathological characteristics of this disease, such as age, sex, degree of differentiation, TNM staging of gastric cancer does not exist (36).
Despite the advances in diagnosis and treatment technologies, the prognosis of gastric cancer patients is still poor, even for those who undergo complete resection of their carcinomas. Having known that DNA methylation is a potentially reversible epigenetic alteration, demethylation inhibitors are thus proposed to be potential new anticancer agents (39,40). Currently, for epigenetic drug therapies tumor suppressor genes are promising targets because many cell cycle inhibitors and tumor suppressor genes are methylated or silenced in cancer cells. The re-expression of tumor suppressor genes is caused by demethylators which lead to their inhibition and apoptosis of cell cycle promotion (41).
In conclusion from the results of this study can be concluded that promoter methylation of Casp8 gene is a frequent epigenetic event in gastric cancer. The results indicated that hypermethylation of this gene was involved in some clinical and pathogenesis of the disease. With few exceptions, the gender, note and tissue type of cancer, correlation has been observed. In contrast the percentage of CpG methylation of promoter region about Apaf1 gene was not in relationship with gastric cancer. But between methylation and pathology information such as stage and grade with few exceptions significant relationship has been observed. Furthermore the methylation pattern of these genes in blood samples, emphasize that epigenetic events have the potential to be as a molecular marker for cancer and has diagnosis and prognostic value for early carcinogenesis detection of gastric cancer.
Acknowledgements
This work was supported by the Biology Research Center of Islamic Azad University, Zanjan, Iran. We would like to express our appreciation to Imam Khomeini and Imam Hussein hospitals for their collaboration. We thank the patients for their participation.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study was conducted at the Biological Research Center of Azad Islamic University of Zanjan and approved by the faculty of medical sciences Ethics Committee (No. IR.IAU.ZANJAN.REC.1396.58). Informed consent was taken from all the patients before entering the study and all the obtained information’s from each participant was completely confidential.
References
- Huang B, Sun Z, Wang Z, et al. Factors associated with peritoneal metastasis in non-serosa-invasive gastric cancer: a retrospective study of a prospectively-collected database. BMC Cancer 2013;13:57. [Crossref] [PubMed]
- Wu CW, Lo SS, Shen KH, et al. Incidence and factors associated with recurrence patterns after intended curative surgery for gastric cancer. World J Surg 2003;27:153-8. [PubMed]
- Inoue M, Tsugane S. Epidemiology of gastric cancer in Japan. Postgrad Med J 2005;81:419-24. [Crossref] [PubMed]
- Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74-108. [Crossref] [PubMed]
- Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol 2006;12:354-62. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Nardone G, Morgner A. Helicobacter pylori and gastric malignancies. Helicobacter 2003;8 Suppl 1:44-52. [Crossref] [PubMed]
- Shibata D, Weiss LM. Epstein-Barr virus-associated gastric adenocarcinoma. Am J Pathol 1992;140:769-74. [PubMed]
- Zang ZJ, Cutcutache I, Poon SL, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet 2012;44:570-4. [Crossref] [PubMed]
- Shi J, Yao D, Liu W, et al. Frequent gene amplification predicts poor prognosis in gastric cancer. Int J Mol Sci 2012;13:4714-26. [Crossref] [PubMed]
- Ushijima T, Asada K. Aberrant DNA methylation in contrast with mutations. Cancer Sci 2010;101:300-5. [Crossref] [PubMed]
- Ziogas D, Roukos D. Epigenetics in gastric cancer: challenges for clinical implications. Ann Surg Oncol 2009;16:2077-8. [Crossref] [PubMed]
- Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res 2001;29:3784-95. [Crossref] [PubMed]
- Kouzarides T. Chromatin modifications and their function. Cell 2007;128:693-705. [Crossref] [PubMed]
- Liang G, Lin JC, Wei V, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A 2004;101:7357-62. [Crossref] [PubMed]
- Momparler RL. Cancer epigenetics. Oncogene 2003;22:6479-83. [Crossref] [PubMed]
- Mitsunaga M, Tsubota A, Nariai K, et al. Early apoptosis and cell death induced by ATX-S10Na (II)-mediated photodynamic therapy are Bax- and p53-dependent in human colon cancer cells. World J Gastroenterol 2007;13:692-8. [Crossref] [PubMed]
- Chiang CT, Way TD, Lin JK. Sensitizing HER2-overexpressing cancer cells to luteolin-induced apoptosis through suppressing p21(WAF1/CIP1) expression with rapamycin. Mol Cancer Ther 2007;6:2127-38. [Crossref] [PubMed]
- Meier P, Finch A, Evan G. Apoptosis in development. Nature 2000;407:796-801. [Crossref] [PubMed]
- Opferman JT, Korsmeyer SJ. Apoptosis in the development and maintenance of the immune system. Nat Immunol 2003;4:410-5. [Crossref] [PubMed]
- Kluck RM, Bossy-Wetzel E, Green DR, et al. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science 1997;275:1132-6. [Crossref] [PubMed]
- Hsu H, Huang J, Shu HB, et al. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity 1996;4:387-96. [Crossref] [PubMed]
- Hsu H, Shu HB, Pan MG, et al. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 1996;84:299-308. [Crossref] [PubMed]
- Kischkel FC, Hellbardt S, Behrmann I, et al. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J 1995;14:5579-88. [PubMed]
- Srinivasula SM, Ahmad M, Fernandes-Alnemri T, et al. Molecular ordering of the Fas-apoptotic pathway: the Fas/APO-1 protease Mch5 is a CrmA-inhibitable protease that activates multiple Ced-3/ICE-like cysteine proteases. Proc Natl Acad Sci U S A 1996;93:14486-91. [Crossref] [PubMed]
- Yang J, Liu X, Bhalla K, et al. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science 1997;275:1129-32. [Crossref] [PubMed]
- Zou H, Henzel WJ, Liu X, et al. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 1997;90:405-13. [Crossref] [PubMed]
- Li P, Nijhawan D, Budihardjo I, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 1997;91:479-89. [Crossref] [PubMed]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415-28. [Crossref] [PubMed]
- Jones PA. The DNA methylation paradox. Trends Genet 1999;15:34-7. [Crossref] [PubMed]
- Singal R, Wang SZ, Sargent T, et al. Methylation of promoter proximal-transcribed sequences of an embryonic globin gene inhibits transcription in primary erythroid cells and promotes formation of a cell type-specific methyl cytosine binding complex. J Biol Chem 2002;277:1897-905. [Crossref] [PubMed]
- Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet 2012;13:97-109. [Crossref] [PubMed]
- Fu W-N, Bertoni F, Kelsey SM, et al. Role of DNA methylation in the suppression of Apaf-1 protein in human leukaemia. Oncogene 2003;22:451-5. [Crossref] [PubMed]
- Wang HL, Bai H, Li Y, et al. Rationales for expression and altered expression of apoptotic protease activating factor-1 gene in gastric cancer. World J Gastroenterol 2007;13:5060-4. [Crossref] [PubMed]
- Huang DF, Fu WN, Shang C, et al. Expression and promoter methylation of Apaf-1 gene in laryngeal squamous cell carcinoma. Yi Chuan Xue Bao 2004;31:1327-31. [PubMed]
- Kupčinskaitė-Noreikienė R, Skieceviciene J, Jonaitis L, et al. CpG island methylation of the MLH1, MGMT, DAPK, and CASP8 genes in cancerous and adjacent noncancerous stomach tissues. Medicina 2013;49:361-6. [PubMed]
- Skiriute D, Vaitkiene P, Saferis V, et al. MGMT, GATA6, CD81, DR4, and CASP8 gene promoter methylation in glioblastoma. BMC Cancer 2012;12:218. [Crossref] [PubMed]
- Kordi Tamandani DM, Sobti RC, Shekari M, et al. CpG island methylation of TMS1/ASC and CASP8 genes in cervical cancer. Eur J Med Res 2009;14:71-5. [Crossref] [PubMed]
- Sarkar S, Goldgar S, Byler S, et al. Demethylation and re-expression of epigenetically silenced tumor suppressor genes: sensitization of cancer cells by combination therapy. Epigenomics 2013;5:87-94. [Crossref] [PubMed]
- Lewandowska J, Bartoszek A. DNA methylation in cancer development, diagnosis and therapy--multiple opportunities for genotoxic agents to act as methylome disruptors or remediators. Mutagenesis 2011;26:475-87. [Crossref] [PubMed]
- Yang J, Wei X, Wu Q, et al. Clinical significance of the expression of DNA methyltransferase proteins in gastric cancer. Mol Med Rep 2011;4:1139-43. [PubMed]


