Potential actionable targets in appendiceal cancer detected by immunohistochemistry, fluorescent in situ hybridization, and mutational analysis
Introduction
Cancers arising from the appendix are rare. In reports of appendectomy specimens, the incidence of malignancy has been reported to be between 0.58% to 0.9% (1,2). The histological spectrum is quite varied, with recent reports through large database studies indicating adenocarcinoma being the most common subtype (3,4). Other histological variants include carcinoid or neuroendocrine and a mixed histology tumor of both carcinoid and adenocarcinoma subtype termed goblet cell carcinoma (5). Finally, another epithelial variant of appendiceal cancer is pseudomyxoma peritonei (PMP), a mucinous neoplasm that clinically presents as gelatinous ascites (6,7). Due to the rarity of these malignancies limited prospective trials exist guiding management. In general, treatment involves surgery for all histological subtypes. For patients with carcinoid appendiceal tumors and goblet cell, appendectomy may be considered in lesions less than 2 cm in size. For lesions 2 cm or more or those with higher grade carcinoid or lymph node involvement right colectomy is indicated (8). For patients with appendiceal adenocarcinoma (<2 cm in size), there is great debate on pursuing simple appendectomies versus hemicolectomy and frequently depends upon opinion (9-11). In patients with PMP, standard treatment involves repeated surgical debulking for symptomatic disease (12). Some clinicians also add intraperitoneal hyperthermia (IPH) treatment or intraperitoneal hyperthermic chemotherapy (IPHC) (13-15).
Outside of localized treatment interventions for appendiceal malignancies, treatment options tend to follow treatment for colorectal malignancies with limited clinical trial data. A recent report from a single center, observational study used FOLFOX-4 as treatment for unresectable or relapsed PMP (7). Furthermore, MGMT methylation was assessed in 42% of these patients, indicating a potential response benefit to temozolomide (7). For carcinoid tumors of the appendix, guidelines recommend treatment to other gastrointestinal neuroendocrine tumors (16). A recent multi-institutional review confirmed current consensus to follow colorectal cancer treatment for appendiceal adenocarcinomas (17). We conducted an analysis of 588 patients with a diagnosis of appendiceal cancer across various histological subtypes who underwent molecular profiling through Caris Life Sciences to look for potential actionable targets and combinations for therapy. Molecular profiling has been used effectively in other cancers to identify novel treatment options. Only two studies to date have reviewed the molecular profile of appendiceal cancers; those studies had very few patients (n=38 and 149) (18,19), and the analysis was limited to gene mutations. Our analysis encompassed multiple profiling platforms from gene alterations to gene amplification and protein expression levels, and 588 patients were evaluated, almost four times more than the previous studies.
Methods
Materials
Data for the 588 specimens with appendix primary malignancies profiled on at least one platform by Caris Life Sciences from 2006 through 2014 were included. Formalin fixed paraffin-embedded (FFPE) samples were sent for analysis from treating physicians around the world (59 countries). The specific histology was extracted from paperwork submitted by the treating physician. Tumors were initially verified by a board certified pathologist for sufficient tumor presence and to confirm the histology. Samples were subsequently analyzed using one or more of the profiling platforms as described below. Biomarkers for analysis varied by case, dependent on tissue availability, physician preference, technology standards over the course of the study, and their potential to be targeted therapeutically and/or based on clinical evidence of a utility in other solid tumors. Carcinosarcomas and adenomas were excluded from analysis. No clinical data, with the exception of basic demographics, were available for this analysis. Biomarkers for analysis were selected based on their potential to be targeted therapeutically and/or based on clinical evidence of a utility in other solid tumors. In accordance with institutional IRB guidelines, because patient identity protection was maintained throughout the study and involves the collection of existing data, the study was considered IRB exempt.
Immunohistochemistry (IHC)
Protein expression was determined by IHC analysis, using commercially available detection kits and automated staining techniques (Benchmark XT, Ventana, and Autostainer Link 48, Dako). Antibodies used included: androgen receptor (AR), topoisomerases 1 and 2 (TOPO1, TOPO2A) (Leica Biosystems); estrogen receptor (ER), progesterone receptor (PR), cMET, Human Epidermal Growth Factor Receptor 2 (HER2) (Ventana); cKIT, epidermal growth factor receptor (EGFR), phosphatase and tensin homolog (PTEN) (Dako), O(6)-methylguanine-methyltransferase (MGMT), P-glycoprotein (PGP), thymidylate synthase (TS) (Invitrogen); transducin-like enhancer of split 3 (TLE3, Santa Cruz); ribonucleotide reductase M1 (RRM1, Protein Tech); SPARC (monoclonal, R&D Systems; polyclonal, Exalpha), tubulin beta-3 chain (TUBB3) (Covance), Excision Repair Cross-Complementation Group 1 (ERCC1, (Abcam), platelet derived growth factor receptor alpha (PDGFRA, Thermo), Programmed cell death protein 1 (PD-1) and Programmed death-ligand 1 (PD-L1) (BD Pharmingen and R&D Systems). IHC thresholds previously validated in other cancers were used, as previously described (20), as cutoffs are not established in appendiceal cancers.
In situ hybridization
Fluorescent in situ hybridization (FISH) was used for evaluation of the HER-2/neu (HER-2/CEP17 probe; HER-2/CEP17 ratio >2.2 was considered amplified), EGFR (EGFR/CEP7 probe EGFR/CEP7 ratio ≥2, or ≥15 EGFR copies per cell in ≥10% of analyzed cells was considered amplified), TOP2A (TOP2/CEP17 probe; TOP2A/CEP17 ratio ≥2.0 was considered amplified), cMET (cMET/CEP7 probe; cMET/CEP7 ratio ≥5 was considered amplified). HER-2/neu and cMET status were more recently evaluated by chromogenic in situ hybridization (INFORM HER-2 Dual ISH DNA Probe Cocktail; commercially available cMET and chromosome 7 DIG probe; Ventana), and used the same scoring system as for FISH.
Mutational analysis
Sanger sequencing
Prior to the availability of CLIA certified NGS, mutation analysis by Sanger sequencing included selected regions of BRAF, KRAS, cKIT, and EGFR genes and was performed by using M13-linked PCR primers designed to amplify targeted sequences. PCR products were bi-directionally sequenced using the BigDye Terminator v1.1 chemistry, analyzed using the 3730 DNA Analyzer (Applied Biosystems). Sequence traces were analyzed using Mutation Surveyor software v3.25 (Soft Genetics).
Next generation sequencing (NGS)
Direct sequence analysis was performed on genomic DNA isolated from FFPE tumor samples using the Illumina MiSeq platform. Average sequencing depth was >1,000×. Specific regions of 47 genes were amplified using the Illumina TruSeq Amplicon Cancer Hotspot panel.
Complete information on thresholds and specific reagents are available at: http://www.carismolecularintelligence.com (20). The variant call was based on nomenclature defined by the ACMGG. Mutations were defined as clinically actionable if the mutation was one for which there is an approved agent available to target, even if the agent is approved for a different tumor type, as well as any clinical trial based on that alteration.
Statistical analysis
The patient population and profiling data were characterized using standard descriptive statistics. When comparing data across the subtypes, groups with less than five cases were not considered. For chemotherapy protein biomarkers, overexpression or loss in at least 60% of samples in a particular subtype were considered clinically significant (mean selected as cutoff).
Results
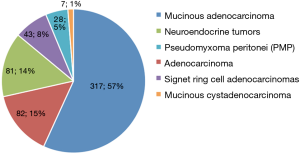
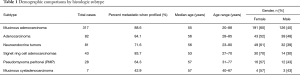
The cases were categorized into histologic subtypes, utilizing information in pathology reports and subsequent pathology review of H&E slides, prior to analysis of molecular patterns in the test results. The majority of cases were adenocarcinomas, at 57% [317] of the total cases (Figure 1). Basic demographic comparisons identified similarities and differences in gender and age distribution by subtypes (Table 1). Specific differences by technology are described by section, followed by an evaluation of the overall differences identified between subtypes.
Full table
Protein expression
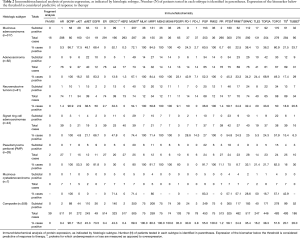
Expression of proteins varied by subtype (Table 2). Specific patterns included overexpression of epidermal growth factor receptor (EGFR) in 74% of cases, with a predominance of adenocarcinomas and PMP’s. TOPO2A was overexpressed on an average in 38% of cases, with the highest rate of in adenocarcinomas (69%). Mismatch repair proteins, MLH1, MSH2, MSH6, and PMS2, were overexpressed in 100% of 75 cases tested. No microsatellite instability was identified in 39 cases tested across subtypes. PR was overexpressed most frequently in PMP (11%) while AR and ER were only aberrated in 2 of >500 cases tested. Other identified aberrations in protein expression included the over-expression of PD-L1 or PD-1 tumor infiltrating lymphocytes in 3% and 31% of cases, respectively and overexpression of cKIT in 18% of mucinous, 16% of adenocarcinoma, and 53% of pseudomyxoma tissues.
Full table
In situ hybridization
A single neuroendocrine case was identified with an increase in EGFR copy number, out of 43 cases tested. No amplifications were identified in TOP2A (n=24), HER2 (n=246), or cMET (n=194) (data not shown).
Gene sequencing
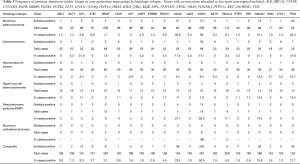
Patterns in a majority of the genomic alterations were different across subtypes. The adenocarcinomas had higher frequency of alterations in APC, BRAF, BRCA2, SMAD4, and TP53 compared to all other subtypes (Table 3), while PMP had the highest incidence of GNAS and KRAS alterations. Alterations in the PI3 kinase pathway (AKT1, PIK3CA, and PTEN) were infrequently identified across subtypes, with 11% PIK3CA alterations in PMP. BRCA2 mutations were identified in 3 of 8 adenocarcinomas tested. Notably, other targetable mutations were found in individual cases, including three cKIT mutations (two mucinous and one neuroendocrine), an ERBB2 mutation, and a BRCA1 mutation.
Full table
Overall differences were observed between the subtypes, in both patterns of gene mutations and protein expression levels. The mucinous adenocarcinoma, adenocarcinoma, and PMP were tumors more likely to exhibit KRAS and GNAS mutations (65%, 47%, and 83%, respectively, compared to only 9% in neuroendocrine and 7% in signet ring cell adenocarcinomas). Notably all subtypes, except for PMP, harbored TP53 mutations. Only mucinous adenocarcinomas harbored ATM mutations, and only mucinous adenocarcinomas and adenocarcinomas harbored BRCA2 mutations (a limited number of cases were tested). APC mutations were found at a significantly higher percent in adenocarcinomas, compared to all other subtypes (P<0.001), and FBXW7 was found between 5–10% in three subtypes (Mucinous, adenocarcinoma, and signet ring) and never in two subtypes (neuroendocrine and PMP). While SMAD4 was found at 22% in adenocarcinomas, it was identified at >10% in all subtypes. BRAF mutations were identified in 8% of adenocarcinomas, in 2% of mucinous adenocarcinomas and not identified in any other subtypes.
Discussion
Molecular profiling of appendiceal cancers suggests a number of treatment options. Treatments based upon the IHC expression of these proteins in appendiceal adenocarcinomas include the use of 5-FU (low TS) that may be combined with irinotecan (due to high TOPO1). Gemcitabine (low RRM1) along with taxanes such as paclitaxel, albumin-bound paclitaxel, docetaxel (low TUBB3, high TLE3) may also be considered as a therapeutic option for appendiceal adenocarcinomas. Due to the low expression ERCC1 seen in appendiceal adenocarcinomas, platinum therapeutics such as cisplatin or oxaliplatin may be combined with either 5-FU or gemcitabine. The adenocarcinomas have a consistent pattern of expression by IHC. In examining mutations in appendiceal adenocarcinoma, KRAS was the most frequent mutation in the mucinous and colonic type adenocarcinoma specimens (85–100% vs. a CRC incidence of 30–50%). Of note was the relative frequency of BRCA2 mutations at 37.5% in the eight colonic type appendiceal adenocarcinomas tested. These mutations may give rise for several different therapeutic treatment options, including those utilizing platinum based therapy (21) and PARP (poly (ADP-ribose) polymerase) inhibition (22). Another difference seen in Figure 2 between colonic-type appendiceal adenocarcinoma and mucinous and signet ring cell is the high mutation rate in APC, a protein involved in the WNT pathway and is seen in individuals with colon cancer and has become a source of agents targeting this pathway in clinical trials (23).
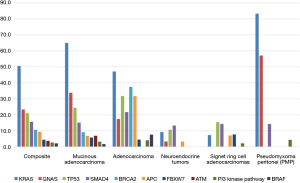
In the neuroendocrine class of appendiceal malignancies, the low expression of and MGMT and TS by IHC can be used as a basis to consider the combination of capecitabine and temozolomide, which has been shown to be beneficial in individuals with neuroendocrine tumors (24). The appendiceal neuroendocrine tumors may also be sensitive to gemcitabine (low RRM1) along with taxanes (low TUBB3, high TLE3). Hot spot mutation analysis yielded few mutations above 10% of specimens with the greatest being SMAD4 and TP53, which currently do not have FDA approved treatments targeting these mutations.
PMP tumor analysis showed low expression of ERCC1 and RRM1 which gemcitabine and cisplatin may be considered if localized therapy is not an option. The low expression of TS also points to the consideration of 5FU and oxaliplatin as an option for therapy. Combination therapy with 5-FU and irinotecan may also be a consideration due to the high expression of TOPO1 in these tumors. Of note, there was no positive expression of either PD-1 or PD-L1 in these tumors along with 100% expression of the mismatch-repair proteins in the samples analyzed. PMP tumor mutations were significant in the presence of KRAS (83.3%), GNAS (57.1%), SMAD4 (14.3%) and PI3KCA (11.1%) mutations, both of which do not have approved targeted therapy options outside of a clinical trial.
Microsatellite instability, seen in 15% of colorectal cancers (25) was not seen in appendiceal cancers. The mismatch repair markers, MLH1, MSH2, MSH6, and PMS2 were overexpressed in 100% of appendiceal cancers tested across subtypes. A recent study showed benefit in checkpoint immune blockade in individuals whose tumors were mismatch-repair deficient (26). Despite the presence of PD-1 and PD-L1 expression seen in appendiceal adenocarcinomas, the normal expression of mismatch-repair proteins points against consideration of immune checkpoint blockade.
The high incidence of cKIT overexpression and number of cKIT mutations across appendiceal cancers, would suggest that treatment with anti cKIT therapies, such as imatinib, may be beneficial in appendiceal cancers, as has been shown in other rare cancers (27).
Conclusions
Recent advances in therapeutics developed to target aberrations in known oncogenic genes have revolutionized medical oncology. Recognition of patterns in biomarker aberrations in different cancers is informing changes in guidelines. Analysis of 588 tumor samples from patients with appendiceal cancer, identified genomic alteration patterns, patterns in gene expression, and aberrations in protein expression that distinguish appendiceal cancers from colorectal cancers and distinguish subtypes of appendiceal cancers. Patterns of genomic alterations in APC, GNAS, and SMAD4 in the neuroendocrine tumors most closely resembled alterations documented in pancreatic adenocarcinomas (28), with the exception of KRAS alterations being significantly lower). The incidence of KRAS alterations in PMP was most similar to rates in pancreatic cancers, while the incidence of APC and KRAS alterations in adenocarcinomas were similar to those in colorectal cancers. Identification of 37% of cases (n=8) with a BRCA2 mutation suggests that a subset of appendiceal adenocarcinomas may have a familial predisposition. These data suggest new avenues for molecularly directed therapies in appendiceal cancers. Overall, this study identified actionable alterations in 99% of cases tested. As knowledge increases of oncogenic pathways and more targeted therapies are approved, continuing to personalize treatment based on the patient’s unique molecular profile will improve the outcomes.
Acknowledgements
None.
Footnote
Conflicts of Interest: J Kimbrough: Employee Caris Life Sciences. The other authors have no conflicts of interest to declare.
Ethical Statement: In accordance with institutional IRB guidelines, because patient identity protection was maintained throughout the study and involves the collection of existing data, the study was considered IRB exempt.
References
- Connor SJ, Hanna GB, Frizelle FA. Appendiceal tumors: retrospective clinicopathologic analysis of appendiceal tumors from 7,970 appendectomies. Dis Colon Rectum 1998;41:75-80. [Crossref] [PubMed]
- Esmer-Sánchez DD, Martínez-Ordaz JL, Román-Zepeda P, et al. Appendiceal tumors. Clinicopathologic review of 5,307 appendectomies. Cir Cir 2004;72:375-8. [PubMed]
- Benedix F, Reimer A, Gastinger I, et al. Study Group Colon/Rectum Carcinoma Primary Tumor. Primary appendiceal carcinoma--epidemiology, surgery and survival: results of a German multi-center study. Eur J Surg Oncol 2010;36:763-71. [Crossref] [PubMed]
- Turaga KK, Pappas SG, Gamblin T. Importance of histologic subtype in the staging of appendiceal tumors. Ann Surg Oncol 2012;19:1379-85. [Crossref] [PubMed]
- Holt N, Grønbæk H. Goblet cell carcinoids of the appendix. ScientificWorldJournal 2013;2013:543696.
- Tang LH. Epithelial neoplasms of the appendix. Arch Pathol Lab Med 2010;134:1612-20. [PubMed]
- Pietrantonio F, Maggi C, Fanetti G, et al. FOLFOX-4 chemotherapy for patients with unresectable or relapsed peritoneal pseudomyxoma. Oncologist 2014;19:845-50. [Crossref] [PubMed]
- Fornaro R, Frascio M, Sticchi C, et al. Appendectomy or right hemicolectomy in the treatment of appendiceal carcinoid tumors? Tumori 2007;93:587-90. [PubMed]
- González-Moreno S, Sugarbaker PH. Right hemicolectomy does not confer a survival advantage in patients with mucinous carcinoma of the appendix and peritoneal seeding. Br J Surg 2004;91:304-11. [Crossref] [PubMed]
- Turaga KK, Pappas S, Gamblin TC. Right hemicolectomy for mucinous adenocarcinoma of the appendix: just right or too much? Ann Surg Oncol 2013;20:1063-7. [Crossref] [PubMed]
- Hata K, Tanaka N, Nomura Y, et al. Early appendiceal adenocarcinoma. A review of the literature with special reference to optimal surgical procedures. J Gastroenterol 2002;37:210-4. [Crossref] [PubMed]
- Hinson FL, Ambrose NS. Pseudomyxoma peritonei. Br J Surg 1998;85:1332-9. [Crossref] [PubMed]
- Hsu KH, Chou CY, Chang YC. Intraperitoneal hyperthermia in the management of pseudomyxoma peritonei. Hepatogastroenterology 2007;54:47-52. [PubMed]
- Sugarbaker PH. Managing the peritoneal surface component of gastrointestinal cancer. Part 1. Patterns of dissemination and treatment options. Oncology (Williston Park) 2004;18:51-9. [PubMed]
- Sugarbaker PH. Managing the peritoneal surface component of gastrointestinal cancer. Part 2. Perioperative intraperitoneal chemotherapy. Oncology (Williston Park) 2004;18:207-19. [PubMed]
- Boudreaux JP, Klimstra DS, Hassan MM, et al. The NANETS consensus guideline for the diagnosis and management of neuroendocrine tumors: well-differentiated neuroendocrine tumors of the Jejunum, Ileum, Appendix, and Cecum. Pancreas 2010;39:753-66. [Crossref] [PubMed]
- Tejani MA, ter Veer A, Milne D, et al. Systemic therapy for advanced appendiceal adenocarcinoma: an analysis from the NCCN Oncology Outcomes Database for colorectal cancer. J Natl Compr Canc Netw 2014;12:1123-30. [PubMed]
- Liu X, Mody K, de Abreu FB, et al. Molecular profiling of appendiceal epithelial tumors using massively parallel sequencing to identify somatic mutations. Clin Chem 2014;60:1004-11. [Crossref] [PubMed]
- Raghav KP, Shetty AV, Kazmi SM, et al. Impact of molecular alterations and targeted therapy in appendiceal adenocarcinomas. Oncologist 2013;18:1270-7. [Crossref] [PubMed]
- Millis SZ, Bryant D, Basu G, et al. Molecular profiling of infiltrating urothelial carcinoma of bladder and nonbladder origin. Clin Genitourin Cancer 2015;13:e37-49. [Crossref] [PubMed]
- Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495-501. [Crossref] [PubMed]
- Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015;33:244-50. [Crossref] [PubMed]
- Laken SJ, Petersen GM, Gruber SB, et al. Familial colorectal cancer in Ashkenazim due to a hypermutable tract in APC. Nat Genet 1997;17:79-83. [Crossref] [PubMed]
- Fine RL, Gulati AP, Krantz BA, et al. Capecitabine and temozolomide (CAPTEM) for metastatic, well-differentiated neuroendocrine cancers: The Pancreas Center at Columbia University experience. Cancer Chemother Pharmacol 2013;71:663-70. [Crossref] [PubMed]
- Poynter JN, Haile RW, Siegmund KD, et al. Associations between smoking, alcohol consumption, and colorectal cancer, overall and by tumor microsatellite instability status. Cancer Epidemiol Biomarkers Prev 2009;18:2745-50. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Perkins J, Boland P, Cohen SJ, et al. Successful imatinib therapy for neuroendocrine carcinoma with activating Kit mutation: a case study. J Natl Compr Canc Netw 2014;12:847-52. [PubMed]
- Millis SZ, Arguello D, Lowery MA, et al. Multiplatform molecular profiling of 2,400 pancreatic adenocarcinomas to identify targets for therapeutic intent. J Clin Oncol 2014;32:abstr 4136.