Current companion diagnostics in advanced colorectal cancer; getting a bigger and better piece of the pie
Introduction
With an ever expanding armamentarium of molecularly targeted therapies that provide clinical benefit in a small subset of patients, the use of predictive biomarkers to appropriately select those who will benefit is crucial. Though the characterization of some predictive markers entails simple testing that can be completed in most labs, such as immunohistochemistry (IHC), other testing can be much more complex (1). The United States Food and Drug Administration (FDA) has recognized the importance of companion diagnostic testing and in 2014 released a position statement highlighting the need for the development and approval of companion in vitro diagnostic tests contemporaneously in order to ensure FDA approval of novel agents (2).
While the pairing of targeted therapies with biomarkers provides an opportunity for personalized care, it also presents significant challenges. As more complex biomarkers emerge, significant variation in assay methodology leads to difficulties in standardizing diagnostic tests and their results. Institutions and funding agencies often struggle with choosing the most cost efficient but clinically useful test from a myriad of competing platforms. The rapid advancement in molecular diagnostics has also resulted in many assays becoming obsolete shortly after adoption as they are replaced with more sensitive or comprehensive tests. Despite these challenges, recent advances have resulted in companion diagnostics with improved clinical performance. Here we review currently available and investigational molecular pathology assays of importance to the treatment of metastatic colorectal cancer (mCRC) and provide insights into future opportunities. We will focus on biomarkers with current or anticipated future actionability (see Figure 1).
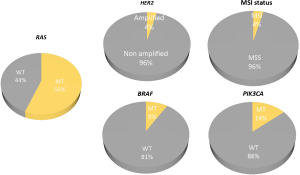
Current standard of care companion diagnostic tests
KRAS/NRAS
Overview and clinical significance
In mCRC, the use of monoclonal antibodies (cetuximab and panitumumab) to target the epidermal growth factor receptor (EGFR) has proven to be an effective treatment strategy (3-9). These agents bind the extra-cellular domain of EGFR and prevent downstream signaling. Correlative studies identified Kirsten rat sarcoma viral oncogene (KRAS) mutations as an important driver of tumor resistance (9). Although initially only KRAS exon 2 (codons 12 and 13) mutations were evaluated as a biomarker for anti-EGFR resistance, recent data has shown that mutations in KRAS exon 3 (codons 59 and 61), exon 4 (codons 117 and 146) or neuroblastoma rat sarcoma viral oncogene (NRAS) exon 2 (codons 12 and 13), 3 (codons 59 and 61), and 4 (codons 117 and 146) are also associated with anti-EGFR resistance (10,11). Together these extended RAS mutations are present in up to 56% of mCRC (12). Due to the strength of RAS testing as a predictive biomarker, guidelines suggest that all patients with mCRC should have extended RAS mutation testing and must not have mutations in RAS if receiving EGFR directed therapy (13-15).
Given the importance to RAS testing, optimizing molecular detection of mutations is of utmost importance. At present, no one methodology is preferred and all assays appear to have similar cost implications in large health care systems (16). Laboratories must either validate their own independent test or adopt a commercially available kit. Concordance between primary tumors and metastatic lesions is high, ranging between 90% and 100% in most studies, suggesting that testing can be completed on whichever lesion is easiest to biopsy, or on archival tissue (17-22). While highly conserved between liver metastasis and primary, some studies have suggested a higher level of discordance (up to 32%) between primary and lung or lymph node metastasis, which may complicate mutation analysis in patients with metastases beyond the liver (23,24). Most samples assessed for RAS status will be formalin-fixed paraffin embedded archival tissue, however in patients without sufficient tissue, cytological samples have been shown to be sufficient for determining RAS and BRAF (v-raf murine sarcoma viral oncogene B1) status with high concordance to primary lesions (17,19,25-27).
While conventional assays identify patients as RAS mutant when a mutation is detected in >10% of reads sequenced, sensitivities approaching 0.1% are now possible (28). This heightened sensitivity allows for greater characterization of sub-clonal populations that may result in early treatment resistance. Multiple studies have described early treatment failure and lack of benefit from EGFR directed therapy in patients with low frequency RAS mutations that would not be detected using standard clinical assays (29-32). It remains to be seen, however, whether these higher sensitivity assays will result in improved patient selection and outcomes. Recently drafted guidelines recommend that assay sensitivity should be at least 5%, although the outcome data supporting this recommendation is limited. Important considerations during assay selection include workload, turnaround time, equipment costs, assay costs, sensitivity and comprehensiveness of assays. Table 1 summarizes some key differences between available platforms. While some assays may allow for detection of only targeted “hot-spot” mutations, others are much more comprehensive and may detect mutations of unclear significance. Sanger sequencing is the gold standard and most techniques are compared to this, however, there are many options available.
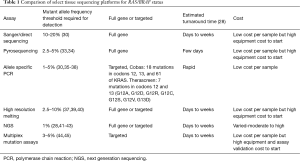
Full table
Direct sequencing techniques
Sanger sequencing and pyrosequencing are methods of direct DNA sequencing which use detection of fluorescent nucleotides or photon emission during nucleotide incorporation to elicit DNA sequence information and provide information about an entire sequenced region (46,47). Direct sequencing is able to determine mutations at all base-pairs throughout an entire gene, but Sanger sequencing has difficulty assessing low frequency mutations or samples with tumor content below 20–30% (48-50). Unlike Sanger sequencing, pyrosequencing allows detection of low level mutations because of its ability to sequence numerous templates concurrently, however it can be limited by the ability to only sequence short templates, which can make detection of uncharacterized mutations more challenging (47,51). Pyrosequencing is more sensitive than Sanger sequencing and comparable in sensitivity to Therascreen, which has been established as a regulatory approved companion biomarker for anti-EGFR inhibitors (33,52). Pyrosequencing is available via independently developed assays or commercially available kits such as PyroMark (Qiagen, Hilden, Germany).
Mutant allele specific polymerase chain reaction (PCR)
Mutant allele specific PCR uses probes for mutated and non-mutated alleles and allows for enrichment of mutant transcripts in samples with low frequency mutations (35). Probes for each allele are labeled with fluorescent reporter dyes that allow detection and quantification. There are a number of modifications to the principle of mutant allele specific PCR that can enhance assay performance, such as peptide-nucleic acid linking or locked nucleic acid incorporation (53). These assays are able to detect mutant allele frequencies that are much lower than Sanger sequencing (as low as 1%) but do not detect mutations outside of those selected for inclusion as primers in the assay (35,36,54). This enhanced sensitivity can result in the re-classification of up to 20% of patients compared to Sanger sequencing or Therascreen and is clinically relevant (30,55). Patients defined as wild-type by PCR based assays have been shown to have improved response rates, progression free, and overall survival (OS) compared to Sanger sequencing defined wild type populations (30,56).
Patented mutant allele specific PCR detection kits, such as the Roche Cobas KRAS mutation kit (Roche Molecular Systems, Inc., Branchburg, NJ, USA) or the Qiagen Therascreen kit (Qiagen, Hilden, Germany), are available and perform well. Cobas uses TaqMelt real-time PCR. Following amplification, samples are heated and mutations are detected by discrepancies between wild type and mutant amplicon melting temperatures. Compared to Sanger sequencing and Therascreen, the Cobas assay has demonstrated superior detection rates, which were in line with pyrosequencing and high resolution melting (HRM) assays (34,37,57). A limitation of the Cobas assay is the inability to confirm which specific mutation is present in a sample. Therascreen uses an allele specific amplification refractory mutation system (ARMS) and incorporates a fluorescent probe (Scorpion) during real-time PCR when a mutation is present in template material. It detects the seven most common mutations in KRAS exon 2 (codon 12 and 13) and has been shown to result in equivalent, if not better sensitivity compared to direct sequencing and HRM, with the ability to detect mutant allele frequencies of 1% (38,49,58,59). The assay identifies the specific mutation which is present in a sample but does not offer extended RAS coverage outside of exon 2 (59).
HRM analysis
HRM analysis uses PCR amplification followed by monitoring for fluorescence changes during heated denaturation to detect mutant alleles. The inclusion of a fluorescent dye or probe that emits more strongly when bound to dsDNA than ssDNA allows detection of variants. Minimum mutant allele frequency required for detection ranges between 2.5% and 10%, making the assay more sensitive than direct sequencing (37,39,40,60). Some reports suggest that HRM RAS detection may result in excessive false positive results but this has not been universally noted and validity was comparable to pyrosequencing and standard sequencing in other studies (59,61). An advantage to HRM is the ability to detect all mutations within multiple genes concurrently (i.e., RAS and BRAF), however the assay is unable to report the specific mutation present in a sample (61).
Gel electrophoresis methods
Single strand conformation polymorphism analysis and denaturing gradient gel electrophoresis both use fluorescent labeling PCR reactions followed by gel electrophoresis with amplified wild-type RAS co-run across gel matrices. Templates with mutations will assume different structures and migrate at different speeds. Mutations are detected by noting when additional bands are present in the gel beyond the wild type band (47,62). Single strand conformation polymorphism analysis, HRM PCR, and Therascreen show similar sensitivities, and appear more sensitive for low frequency mutations than pyrosequencing (62). Gel electrophoresis methods can detect an array of mutations across entire genes but do not identify the specific gene mutation present (63). The Ampli-set-K-RAS commercial kit (Bird, Siena, Italy) uses restriction fragment length polymorphism PCR and has been shown to have similar sensitivity and specificity to Therascreen and multiplex assays (64).
Next generation sequencing (NGS)
NGS uses massively paralleled sequencing technology to perform sequencing with significant depth and covering many genes with similar time and resource costs compared to sequencing a single mutation with Sanger sequencing (65). It can utilize highly selective hot-spot panels with significant depth at each mutation site or more comprehensive sequencing such as whole-exome or whole-genome analysis. With more comprehensive sequencing, data management and informatics pipelines becomes increasingly important to deal with the massive amounts of information generated. The costs of NGS are falling significantly and costs to sequence a single genome are now on the scale of a few thousand dollars, with targeted panels available for several hundred dollars a sample. Both Haley et al. and Altimari et al. demonstrated that NGS had better sensitivity and specificity than Sanger sequencing, pyrosequencing and ARMS-Scorpion PCR assays for the detection of known KRAS mutations (41,42). NGS has a sensitivity as low as 1% mutant allele frequency and had 100% concordance with a panel of KRAS mutant patients compared to 98% with Therascreen (43). Other major benefits of NGS will be discussed later when we address the roll of multiplex panels in mCRC.
Emerging biomarkers in colorectal cancer
BRAF
Overview and clinical significance
BRAFV600E mutations are found in 8–10% of mCRC and are strongly associated with RAS wild-type and microsatellite instable (MSI) tumors (66). BRAF mutant tumors associated with MSI result from high level CpG island hypermethylation [CpG island hypermethylation phenotype (CIMP)+] (67). This CIMP+ phenotype accounts for almost all cases of BRAFV600E mutant metastatic CRC. Testing for BRAFV600E mutation is recommended for its prognostic and potentially predictive role (68). Results of biomarker analysis from the PRIME randomized trial comparing the efficacy of panitumumab plus oxaliplatin, fluorouracil, and leucovorin (FOLFOX4) with FOLFOX4 alone, revealed that the presence of BRAFV600E mutation in RAS wild-type tumors portends poor prognosis, regardless of treatment (4). The median progression-free survival (PFS) and OS in BRAF mutant patients compared to wild type patients was 6.1 vs. 10.8 months and 10.5 vs. 28.3 months, respectively (69). Due to their poor prognosis, these patients need to be identified early and considered for clinical trials assessing BRAF inhibitors (70). Additionally, BRAFV600E mutation is strongly associated with MLH1 promoter methylation and can therefore distinguish between sporadic and hereditary (Lynch-related) MLH1-deficient MSI CRC (71).
BRAFV600E mutation assays
BRAFV600E mutation testing is performed commonly using PCR based methods. In a study comparing PCR-Sequencing (Sanger sequencing) with a real time PCR (allelic discrimination) assay, real time PCR was found to be more sensitive for detection of BRAF mutations in CRC, especially in cases with an allelic frequency of less than 20% (72). NGS platforms can also be used for detecting BRAFV600E mutations and offers the advantage of testing multiple genes for mutations of interest. In a prospective comparison, the performance of the Ion Torrent NGS assay was found comparable to Sanger sequencing and detected similar proportions of BRAF mutant cases (73). Since these methods require DNA extraction, they are resource intensive, and often inaccessible. Notably, BRAFV600E mutation testing can facilitate screening of Lynch syndrome related MSI CRCs (74,75). Emerging data has shown that BRAFV600E mutation in plasma cfDNA can be tested using PCR based tests, has moderate concordance (concordance rate of 74%) with standard tumor tissue testing and may have prognostic and predictive significance (76).
Human epidermal growth factor receptor 2 (HER2)
Overview and clinical significance
HER2 amplifications are found in 3–4% of all CRC and are strongly associated with RAS and BRAF wild-type tumors. In patients harboring wild-type RAS/BRAF genes, HER2 amplifications are seen in 5–6% cases (66,77). Presence of HER2 amplification has been implicated in resistance to anti-EGFR monoclonal antibodies (78). In the clinical setting, HER2 gene copy number status significantly correlated with differential response rate (RR), PFS and OS. Patients with HER2 amplification have shorter PFS on treatment with anti-EGFR based therapies compared to HER2 non-amplified cases (79). HER2 amplification has been validated as a negative predictive biomarker for anti-EGFR antibody therapy in metastatic CRC. Patients with HER2 amplification had significantly shorter PFS (median: 2.9 vs. 8.1 months) compared to HER2 non-amplified patients on anti-EGFR based therapy (80). Furthermore, these patients can derive benefit from dual-anti-HER2 inhibition using trastuzumab in combination with lapatinib (HERACLES study) or trastuzumab and pertuzumab (My Pathway study) and should be referred for clinical trials assessing HER-2 directed therapy (81-83).
HER2 amplification testing
No consensus exists regarding methodology for HER2 amplification testing in CRC. The HERACLES study used HER2 IHC for patient selection (82). Using this cohort, the investigators proposed a criterion for HER2-IHC positivity to identify HER2 amplification in CRC. IHC was performed using both HercepTest antibody and Bench Mark Ultrasystem using the VENTANA 4B5 antibody. In this cohort, none of the IHC 0 or 1+ cases were amplified. Furthermore, there was complete concordance between silver in situ hybridization (SISH) and fluorescent in situ hybridization (FISH) (84). However, in the absence of prospective validation, the application of this algorithmic interpretation of IHC and ISH in CRC is incomplete. In another systematic analysis of 2,573 CRC cases using HER2 IHC and in situ hybridization (ISH), the diagnostic sensitivity and specificity of HER2 IHC was 71% and 96%, respectively. HER2 IHC scores of 0 or 1+ exhibited good agreement with ISH (concordance rate of 97%). However, the concordance rates of HER2 IHC 2+ and 3+ were (38% and 78%, respectively) low, necessitating the additional need for ISH analysis to confirm HER2 status in these cases (85). Although, not validated in CRC, NGS assay can accurately identify HER2 amplifications in breast cancer samples (86). HER2 amplifications in plasma cfDNA can be identified using digital PCR. In mCRC patients who developed resistance to anti-EGFR antibodies, 22% (4/18) of patients were found to have HER2 amplifications (87), although this methodology requires tissue concordance validation studies before it is ready for clinical implementation.
PIK3CA
Overview and clinical significance
Phosphatidylinositol-4,5-bisphosphate 3-kinase (PIK3CA) is a key enzyme in the MTOR signaling pathway. Mutations in exon 9 and 20 have been reported in 14–32%, and had been reported to associate with resistance to EGFR directed therapy (88-91). However, these early studies may have been confounded by the frequent co-mutation of both PIK3CA and KRAS in CRC tumors, and subsequent studies have not demonstrated a role for PIK3CA as an independent predictive biomarker for EGFR antibodies. Exon 20 mutations appear to be more strongly associated with EGFR resistance, however a recent meta-analysis suggests the prognostic significance of PIK3CA mutations is unclear at this point (92,93). PIK3CA mutations have been noted to be a potential biomarker for benefit from ASA or COX-2 inhibition in the adjuvant setting after several studies demonstrated improved outcomes in patients with PIK3CA mutations taking ASA after a colorectal cancer diagnosis (94,95). At this point in time there is no proven therapeutic intervention directed at PIK3CA mutations in mCRC. Due to this, testing for these mutations is not indicated outside of a clinical trial.
Mutation detection
Similar to KRAS and BRAF, PIK3CA mutations are highly conserved between primary and metastasis, with a concordance of over 90% (19). There is no preferred assay to detect PIK3CA mutations and individual reports suggest NGS platforms, gel electrophoresis, and PCR sequencing all have excellent sensitivities, while Sanger sequencing is less sensitive. HRM has been shown to have similar sensitivity to Sanger sequencing, while pyrosequencing appears to be more sensitive than Sanger sequencing (96-100).
Microsatellite instability (MSI)
While MSI is a long standing prognostic biomarker in early stage CRC, interest in this biomarker in the metastatic setting has been renewed with the advent of immune checkpoint inhibitors (101-103). The use of pembrolizumab in MSI-high CRCs has been associated with significant response rates (104). A plausible hypothesis is that deficient DNA repair results in higher mutational burden in MSI-high tumors, and the resultant increase in neo-antigens provides an immunogenic environment. Notably, level of mutational burden correlates with response to immune checkpoint inhibitors in other tumor sites (105,106). MSI is present in 15–20% of all CRCs and about 4% of mCRC and arises from either germline mutations in mismatch repair (MMR) proteins (MSH2, MSH6, MLH1, PMS2) or from somatic hypermethylation of the MLH1 promoter (107,108). Loss of these MMR proteins results in expansions of short repetitive sequences throughout the genome called microsatellites that can be detected via PCR-based assays which target standard DNA sequences containing these repeats. A simpler way to infer MMR function is IHC to detect loss of expression in the four key MMR proteins. IHC has been shown to have similar sensitivity to PCR based MSI testing, however is not able to distinguish germ line mutations from somatic hypermethylation of MLH1 (1). While this deficit has implications for hereditary screening programs, it does not impact the phenotypic behavior of tumors. Besides MSI status, other markers such as tumor infiltrating lymphocytes and mutational burden have not been clinically validated in colorectal cancer.
Novel assay methodologies
Cell free DNA (cfDNA), circulating tumor DNA (ctDNA), and circulating tumor cells
There are a number of novel assays capable of assessing RAS and BRAF status from plasma or serum. The ability to ascertain this information from a “liquid biopsy” reduces complications associated with biopsy procedures and provides another means to assess response to therapy and detect emergence of resistance. ctDNA genotyping of RAS and BRAF has been shown to have similar, if not better, ability to detect low frequency mutations compared to tissue based assays in known RAS/BRAF mutant patients and allows monitoring of clonal dynamics (109,110). While many studies report rates of discordance between 20% and 30% in RAS and nearing 10% in BRAF for cfDNA, ctDNA, and CTC based assays, it is difficult to interpret whether this demonstrates inadequacies in an assay or temporal clonal dynamics (111-115). A recent phase II trial of irinotecan and cetuximab demonstrated that cfDNA KRAS mutation status was often discordant compared to archival tissue, but was actually more predictive of patient response and survival than archival status. This work highlights how plasma assays may actually provide a better representation of the current mutational burden of a patient (116). The ability to follow these clonal dynamics has also been shown to offer potential re-treatment options. Siravegna et al. demonstrated emergence of a RAS resistant clone after treatment that subsequently diminished upon cessation of EGFR directed therapy. These patients were able to be re-challenged with cetuximab/panitumumab and achieved responses prior to the re-emergence of the prior clone (109).
Multigene panels
As we move towards the increasing use of molecularly targeted agents, it is becoming crucial to assess samples for multiple biomarkers concurrently. Utilizing multiple different assays to test for individual targets is challenging due to time constraints, cost, and pathologic specimen exhaustion. At this time, whole genome and whole exome sequencing are still out of reach for clinical utilization, however the use of NGS targeted gene panels allows a focused assessment of key genes and have been adopted at many institutions. By inserting a barcode into sample template material, a multiplexed platform that can sequence multiple samples concurrently is possible, facilitating increased throughput. Once barcoded, oligonucleotides are hybridized to a solid matrix with mutation specific sequences followed by PCR amplification to create clonal clusters. DNA synthesis of sequences complementary to the clonal clusters is detected by a marker such as pH change or fluorescence emission, depending on the platform used, and this information is computationally merged across all of the concurrent reactions to create the full output of the sequencing.
Multiplex assays allow assessment of numerous genes within one reaction and have similar or better sensitivity to direct sequencing methods (58,117,118). They are cheaper than direct sequencing and are better able to deal with degraded or poor quality DNA. Typically, a panel is designed to detect “hot-spot” mutations that have been previously described with clinical significance, however mutation coverage can differ based on the specific assay. The decision concerning how much of a gene or the number of genes to include on a panel must be balanced with the depth of coverage desired. By limiting the size of a panel, increasing depth at each mutation is possible with similar cost. Mutant allele frequency required for detection is reported in the range of 3–5% and with significant depth of coverage or paired reads that span breakpoints, copy number variation (CNV) can be detected with sensitivities nearing 100% (44,45,86,119,120). This does require that the break point and adjacent territory both have adequate coverage which may not always be present depending on the particular panel. Detection of gene fusions is difficult unless the panel is designed to include targets for a known fusion. Most novel fusions are detected using WGS or RNA-Seq technology (121,122).
Between 29–72% of patients had potentially actionable targets after sequencing with multiplex panels in results from several large cohorts (123-126). The number of actionable targets is highly variable depending on the available compounds for treatment at a center and the number of mutations assessed in each panel. At this point in time, these panels are most effective in centers with large early drug development programs that can provide access to numerous investigational compounds. The Assessment of Targeted Therapies Against Colorectal Cancer (ATTACC) umbrella trial protocol at MD Anderson Cancer Center recently reported the results of the first 484 patients screened. Ninety-five percent of patients had a biomarker identified, of which 31.2% were enrolled onto one of the 18 companion trials available during the study (127). Other ongoing umbrella and basket studies, such as NCI-MATCH, will hopefully demonstrate the utility of multigene panels to complete accrual to trials of uncommon molecular subtypes (128).
With advances in NGS technology and reduction in cost, panels with a larger number of genes and/or hotspots are possible. Given the lack of targeted agents that have demonstrated clinical effectiveness in metastatic colorectal cancer at this point in time, there is no clinical indication to utilize larger panels beyond those that would include RAS and potentially BRAF mutations outside of a clinical trial setting. One of the major reasons to include a larger number of genes within a panel is to create a platform that can be used across numerous tumor sites to save resources and allow batching of different histologies together.
Conclusions
Despite a lack of new agents available for the treatment of mCRC over the past decade, the refinement of current biomarkers to better select patients who will benefit from EGFR inhibition has been an important step towards improving outcomes. The current drug development pipeline includes exciting new targeted agents that may be available in the next few year, however these therapies are likely to provide benefit in only a subset of patients. In order for these agents to move into the clinic and receive regulatory approval, they will require tandem development of companion diagnostic tests. Current commonly assessed biomarkers are outlined in Table 2. Selection of the appropriate companion diagnostic in some settings may be dictated by regulatory authorities during drug approval, however in many situations institutions will need to select one of a variety of assays that may provide similar information. These decisions will be driven by assay performance, cost, and labor intensity. With the rapid improvement in NGS technology, significant reduction in sequencing cost, and ability to test specimens from many patients on the same assay, we expect future companion diagnostics will rely heavily on multiplex panels that can effectively screen patients for numerous agents concurrently. These assays not only benefit patients by providing information about multiple mutational targets concurrently, but better utilize pathologic specimens.

Full table
Acknowledgements
JM Loree is a member of the University of British Columbia Clinician Investigator program and was the recipient of the Canadian Association of Medical Oncology 2016 Research Fellowship, the Conquer Cancer Foundation of ASCO-J. Edward Mahoney Foundation Young Investigator Award, and the Royal College of Physicians and Surgeons of Canada Detweiler Travelling Fellowship. We thank these programs for providing their support and making this work possible. This research was performed in partial fulfillment of the requirements for the MS degree from The University of Texas Graduate School of Biomedical Sciences at Houston.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn 2008;10:293-300. [Crossref] [PubMed]
- FDA. In Vitro Companion Diagnostic Devices: Guidance for Industry and Food and Drug Administration Staff. United States Food and Drug Administration. 2014.
- Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 2004;351:337-45. [Crossref] [PubMed]
- Douillard JY, Siena S, Cassidy J, et al. Final results from PRIME: randomized phase III study of panitumumab with FOLFOX4 for first-line treatment of metastatic colorectal cancer. Ann Oncol 2014;25:1346-55. [Crossref] [PubMed]
- Poulin-Costello M, Azoulay L, Van Cutsem E, et al. An analysis of the treatment effect of panitumumab on overall survival from a phase 3, randomized, controlled, multicenter trial (20020408) in patients with chemotherapy refractory metastatic colorectal cancer. Target Oncol 2013;8:127-36. [Crossref] [PubMed]
- Maughan TS, Adams RA, Smith CG, et al. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: Results of the randomised phase 3 MRC COIN trial. Lancet 2011;377:2103-14. [Crossref] [PubMed]
- Van Cutsem E, Lenz HJ, Köhne CH, et al. Fluorouracil, Leucovorin, and Irinotecan Plus Cetuximab Treatment and RAS Mutations in Colorectal Cancer. J Clin Oncol 2015;33:692-700. [Crossref] [PubMed]
- Bokemeyer C, Köhne CH, Ciardiello F, et al. FOLFOX4 plus cetuximab treatment and RAS mutations in colorectal cancer. Eur J Cancer 2015;51:1243-52. [Crossref] [PubMed]
- Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008;359:1757-65. [Crossref] [PubMed]
- Sorich MJ, Wiese MD, Rowland A, et al. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized controlled trials. Ann Oncol 2015;26:13-21. [Crossref] [PubMed]
- Therkildsen C, Bergmann TK, Henrichsen-Schnack T, et al. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis. Acta Oncol 2014;53:852-64. [Crossref] [PubMed]
- Peeters M, Kafatos G, Taylor A, et al. Prevalence of RAS mutations and individual variation patterns among patients with metastatic colorectal cancer: A pooled analysis of randomised controlled trials. Eur J Cancer 2015;51:1704-13. [Crossref] [PubMed]
- Allegra CJ, Rumble RB, Hamilton SR, et al. Extended RAS Gene Mutation Testing in Metastatic Colorectal Carcinoma to Predict Response to Anti-Epidermal Growth Factor Receptor Monoclonal Antibody Therapy: American Society of Clinical Oncology Provisional Clinical Opinion Update 2015. J Clin Oncol 2016;34:179-85. [Crossref] [PubMed]
- National Comprehensive Cancer Network. Colon Cancer - 2016 Clinical Practice Guidelines. Available online: www.nccn.org
- Van Cutsem E, Cervantes A, Nordlinger B, et al. Metastatic colorectal cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 2014;25:iii1-9. [Crossref] [PubMed]
- Westwood M, Van Asselt T, Ramaekers B, et al. KRAS mutation testing of tumours in adults with metastatic colorectal cancer: A systematic review and cost-effectiveness analysis. Health Technol Assess 2014;18:1-132. [Crossref] [PubMed]
- Miglio U, Mezzapelle R, Paganotti A, et al. Mutation analysis of KRAS in primary colorectal cancer and matched metastases by means of highly sensitivity molecular assay. Pathol Res Pract 2013;209:233-6. [Crossref] [PubMed]
- Mariani P, Lae M, Degeorges A, et al. Concordant analysis of KRAS status in primary colon carcinoma and matched metastasis. Anticancer Res 2010;30:4229-35. [PubMed]
- Mao C, Wu XY, Yang ZY, et al. Concordant analysis of KRAS, BRAF, PIK3CA mutations, and PTEN expression between primary colorectal cancer and matched metastases. Sci Rep 2015;5:8065. [Crossref] [PubMed]
- Vignot S, Lefebvre C, Frampton GM, et al. Comparative analysis of primary tumour and matched metastases in colorectal cancer patients: Evaluation of concordance between genomic and transcriptional profiles. Eur J Cancer 2015;51:791-9. [Crossref] [PubMed]
- Baas JM, Krens LL, Guchelaar HJ, et al. Concordance of predictive markers for EGFR inhibitors in primary tumors and metastases in colorectal cancer: a review. Oncologist 2011;16:1239-49. [Crossref] [PubMed]
- Cejas P, López-Gómez M, Aguayo C, et al. Analysis of the concordance in the EGFR pathway status between primary tumors and related metastases of colorectal cancer patients:implications for cancer therapy. Curr Cancer Drug Targets 2012;12:124-31. [Crossref] [PubMed]
- Han CB, Li F, Ma JT, et al. Concordant KRAS mutations in primary and metastatic colorectal cancer tissue specimens: a meta-analysis and systematic review. Cancer Invest 2012;30:741-7. [Crossref] [PubMed]
- Kim MJ, Lee HS, Kim JH, et al. Different metastatic pattern according to the KRAS mutational status and site-specific discordance of KRAS status in patients with colorectal cancer. BMC Cancer 2012;12:347. [Crossref] [PubMed]
- Troncone G, Malapelle U, Cozzolino I, et al. KRAS mutation analysis on cytological specimens of metastatic colo-rectal cancer. Diagn Cytopathol 2010;38:869-73. [Crossref] [PubMed]
- Bozzetti C, Negri FV, Azzoni C, et al. Epidermal growth factor receptor and Kras gene expression: reliability of mutational analysis on cytological samples. Diagn Cytopathol 2013;41:595-8. [Crossref] [PubMed]
- Pang NK, Nga ME, Chin SY, et al. KRAS and BRAF mutation analysis can be reliably performed on aspirated cytological specimens of metastatic colorectal carcinoma. Cytopathology 2011;22:358-64. [Crossref] [PubMed]
- Jimeno A, Messersmith W, Hirsch FR, et al. KRAS mutations and sensitivity to epidermal growth factor receptor inhibitors in colorectal cancer: Practical application of patient selection. J Clin Oncol 2009;27:1130-6. [Crossref] [PubMed]
- Kimura T, Okamoto K, Miyamoto H, et al. Clinical benefit of high-sensitivity KRAS mutation testing in metastatic colorectal cancer treated with anti-EGFR antibody therapy. Oncology 2012;82:298-304. [Crossref] [PubMed]
- Dono M, Massucco C, Chiara S, et al. Low percentage of KRAS mutations revealed by locked nucleic acid polymerase chain reaction: implications for treatment of metastatic colorectal cancer. Mol Med 2013;18:1519-26. [PubMed]
- Molinari F, Felicioni L, Buscarino M, et al. Increased detection sensitivity for KRAS mutations enhances the prediction of anti-EGFR monoclonal antibody resistance in metastatic colorectal cancer. Clin Cancer Res 2011;17:4901-14. [Crossref] [PubMed]
- Tougeron D, Lecomte T, Pagès JC, et al. Effect of low-frequency KRAS mutations on the response to anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol 2013;24:1267-73. [Crossref] [PubMed]
- Sundström M, Edlund K, Lindell M, et al. KRAS analysis in colorectal carcinoma: analytical aspects of Pyrosequencing and allele-specific PCR in clinical practice. BMC Cancer 2010;10:660. [Crossref] [PubMed]
- Gonzalez de Castro D, Angulo B, Gomez B, et al. A comparison of three methods for detecting KRAS mutations in formalin-fixed colorectal cancer specimens. Br J Cancer 2012;107:345-51. [Crossref] [PubMed]
- Lang AH, Drexel H, Geller-Rhomberg S, et al. Optimized allele-specific real-time PCR assays for the detection of common mutations in KRAS and BRAF. J Mol Diagn 2011;13:23-8. [Crossref] [PubMed]
- Lièvre A, Bachet JB, Boige V, et al. KRAS Mutations As an Independent Prognostic Factor in Patients With Advanced Colorectal Cancer Treated With Cetuximab. J Clin Oncol 2008;26:374-9. [Crossref] [PubMed]
- Harlé A, Busser B, Rouyer M, et al. Comparison of COBAS 4800 KRAS, TaqMan PCR and High Resolution Melting PCR assays for the detection of KRAS somatic mutations in formalin-fixed paraffin embedded colorectal carcinomas. Virchows Arch 2013;462:329-35. [Crossref] [PubMed]
- Angulo B, García-García E, Martínez R, et al. A commercial real-time PCR kit provides greater sensitivity than direct sequencing to detect KRAS mutations: a morphology-based approach in colorectal carcinoma. J Mol Diagn 2010;12:292-9. [Crossref] [PubMed]
- Krypuy M, Newnham GM, Thomas DM, et al. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer 2006;6:295. [Crossref] [PubMed]
- Dijkstra JR, Heideman DA, Meijer GA, et al. KRAS mutation analysis on low percentage of colon cancer cells: The importance of quality assurance. Virchows Arch 2013;462:39-46. [Crossref] [PubMed]
- Altimari A, De Biase D, De Maglio G, et al. 454 next generation-sequencing outperforms allele-specific PCR, sanger sequencing, and pyrosequencing for routine KRAS mutation analysis of formalin-fixed, paraffin-embedded samples. Onco Targets Ther 2013;6:1057-64. [PubMed]
- Haley L, Tseng LH, Zheng G, et al. Performance characteristics of next-generation sequencing in clinical mutation detection of colorectal cancers. Mod Pathol 2015;28:1390-9. [Crossref] [PubMed]
- Gao J, Wu H, Wang L, et al. Validation of targeted next-generation sequencing for RAS mutation detection in FFPE colorectal cancer tissues: comparison with Sanger sequencing and ARMS-Scorpion real-time PCR. BMJ Open 2016;6:e009532. [Crossref] [PubMed]
- D’Haene N, Le Mercier M, De Nève N, et al. Clinical validation of targeted next generation sequencing for colon and lung cancers. PLoS One 2015;10:e0138245. [Crossref] [PubMed]
- Heideman DA, Lurkin I, Doeleman M, et al. KRAS and BRAF mutation analysis in routine molecular diagnostics: Comparison of three testing methods on formalin-fixed, paraffin-embedded tumor-derived DNA. J Mol Diagn 2012;14:247-55. [Crossref] [PubMed]
- Dufort S, Richard MJ, de Fraipont F. Pyrosequencing method to detect KRAS mutation in formalin-fixed and paraffin-embedded tumor tissues. Anal Biochem 2009;391:166-8. [Crossref] [PubMed]
- Shackelford RE, Whitling NA, McNab P, et al. KRAS Testing: A Tool for the Implementation of Personalized Medicine. Genes Cancer 2012;3:459-66. [Crossref] [PubMed]
- Gao J, Li YY, Sun PN, et al. Comparative analysis of dideoxy sequencing, the KRAS StripAssay and pyrosequencing for detection of KRAS mutation. World J Gastroenterol 2010;16:4858-64. [Crossref] [PubMed]
- Tol J, Dijkstra JR, Vink-Börger ME, et al. High sensitivity of both sequencing and real-time PCR analysis of KRAS mutations in colorectal cancer tissue. J Cell Mol Med 2010;14:2122-31. [Crossref] [PubMed]
- Malapelle U, Bellevicine C, Salatiello M, et al. Sanger sequencing in routine KRAS testing: a review of 1720 cases from a pathologist’s perspective. J Clin Pathol 2012;65:940-4. [Crossref] [PubMed]
- Weidlich S, Walsh K, Crowther D, et al. Pyrosequencing-based methods reveal marked inter-individual differences in oncogene mutation burden in human colorectal tumours. Br J Cancer 2011;105:246-54. [Crossref] [PubMed]
- Tsiatis AC, Norris-Kirby A, Rich RG, et al. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn 2010;12:425-32. [Crossref] [PubMed]
- Kobunai T, Watanabe T, Yamamoto Y, et al. The frequency of KRAS mutation detection in human colon carcinoma is influenced by the sensitivity of assay methodology: A comparison between direct sequencing and real-time PCR. Biochem Biophys Res Commun 2010;395:158-62. [Crossref] [PubMed]
- Chang YS, Yeh KT, Hsu NC, et al. Detection of N-, H-, and KRAS codons 12, 13, and 61 mutations with universal RAS primer multiplex PCR and N-, H-, and KRAS-specific primer extension. Clin Biochem 2010;43:296-301. [Crossref] [PubMed]
- Efrati E, Elkin H, Peerless Y, et al. LNA-based PCR clamping enrichment assay for the identification of KRAS mutations. Cancer Biomark 2010-2011;8:89-94. [Crossref] [PubMed]
- Shan L, Li M, Ma J, et al. PCR-based assays versus direct sequencing for evaluating the effect of KRAS status on anti-EGFR treatment response in colorectal cancer patients: A systematic review and meta-analysis. PLoS One 2014;9:e107926. [Crossref] [PubMed]
- Lee S, Brophy VH, Cao J, et al. Analytical performance of a PCR assay for the detection of KRAS mutations (codons 12/13 and 61) in formalin-fixed paraffin-embedded tissue samples of colorectal carcinoma. Virchows Arch 2012;460:141-9. [Crossref] [PubMed]
- Bando H, Yoshino T, Shinozaki E, et al. Simultaneous identification of 36 mutations in KRAS codons 61 and 146, BRAF, NRAS, and PIK3CA in a single reaction by multiplex assay kit. BMC Cancer 2013;13:405. [Crossref] [PubMed]
- Franklin WA, Haney J, Sugita M, et al. KRAS mutation: comparison of testing methods and tissue sampling techniques in colon cancer. J Mol Diagn 2010;12:43-50. [Crossref] [PubMed]
- Akiyoshi K, Yamada Y, Honma Y, et al. KRAS mutations in patients with colorectal cancer as detected by high-resolution melting analysis and direct sequencing. Anticancer Res 2013;33:2129-34. [PubMed]
- Borras E, Jurado I, Hernan I, et al. Clinical pharmacogenomic testing of KRAS, BRAF and EGFR mutations by high resolution melting analysis and ultra-deep pyrosequencing. BMC Cancer 2011;11:406. [Crossref] [PubMed]
- Whitehall V, Tran K, Umapathy A, et al. A multicenter blinded study to evaluate KRAS mutation testing methodologies in the clinical setting. J Mol Diagn 2009;11:543-52. [Crossref] [PubMed]
- Hayes VM, Westra JL, Verlind E, et al. New comprehensive denaturing-gradient-gel- electrophoresis assay for KRAS mutation detection applied to paraffin-embedded tumours. Genes Chromosomes Cancer 2000;29:309-14. [Crossref] [PubMed]
- Cavallini A, Valentini AM, Lippolis C, et al. KRAS genotyping as biomarker in colorectal cancer: A comparison of three commercial kits on histologic material. Anticancer Res 2010;30:5251-6. [PubMed]
- Dong L, Wang W, Li A, et al. Clinical Next Generation Sequencing for Precision Medicine in Cancer. Curr Genomics 2015;16:253-63. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [Crossref] [PubMed]
- Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 2006;38:787-93. [Crossref] [PubMed]
- Schmoll HJ, Van Cutsem E, Stein A, et al. Esmo consensus guidelines for management of patients with colon and rectal cancer. A personalized approach to clinical decision making. Ann Oncol 2012;23:2479-516. [Crossref] [PubMed]
- Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369:1023-34. [Crossref] [PubMed]
- Clarke CN, Kopetz ES. BRAF mutant colorectal cancer as a distinct subset of colorectal cancer: clinical characteristics, clinical behavior, and response to targeted therapies. J Gastrointest Oncol 2015;6:660-7. [PubMed]
- Parsons MT, Buchanan DD, Thompson B, et al. Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet 2012;49:151-7. [Crossref] [PubMed]
- Roma C, Rachiglio AM, Pasquale R, et al. BRAF V600E mutation in metastatic colorectal cancer: methods of detection and correlation with clinical and pathologic features. Cancer Biol Ther 2016;17:840-8. [Crossref] [PubMed]
- Malapelle U, Vigliar E, Sgariglia R, et al. Ion Torrent next-generation sequencing for routine identification of clinically relevant mutations in colorectal cancer patients. J Clin Pathol 2015;68:64-8. [Crossref] [PubMed]
- Toon CW, Walsh MD, Chou A, et al. BRAFV600E immunohistochemistry facilitates universal screening of colorectal cancers for Lynch syndrome. Am J Surg Pathol 2013;37:1592-602. [Crossref] [PubMed]
- Boissière-Michot F, Frugier H, Ho-Pun-Cheung A, et al. Immunohistochemical staining for p16 and BRAFV600E is useful to distinguish between sporadic and hereditary (Lynch syndrome-related) microsatellite instable colorectal carcinomas. Virchows Arch 2016;469:135-44. [Crossref] [PubMed]
- Janku F, Claes B, Huang HJ, et al. BRAF mutation testing with a rapid, fully integrated molecular diagnostics system. Oncotarget 2015;6:26886-94. [Crossref] [PubMed]
- Richman SD, Southward K, Chambers P, et al. HER2 overexpression and amplification as a potential therapeutic target in colorectal cancer: analysis of 3256 patients enrolled in the QUASAR, FOCUS and PICCOLO colorectal cancer trials. J Pathol 2016;238:562-70. [Crossref] [PubMed]
- Bertotti A, Migliardi G, Galimi F, et al. A molecularly annotated platform of patient-derived xenografts ("xenopatients") identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov 2011;1:508-23. [Crossref] [PubMed]
- Martin V, Landi L, Molinari F, et al. HER2 gene copy number status may influence clinical efficacy to anti-EGFR monoclonal antibodies in metastatic colorectal cancer patients. Br J Cancer 2013;108:668-75. [Crossref] [PubMed]
- Raghav KP, Overman MJ, Yu R, et al. HER2 amplification as a negative predictive biomarker for anti epidermal growth factor receptor antibody therapy in metastatic colorectal cancer. J Clin Oncol 2016;34:abstr 3517.
- Treating Tumors by Molecular Profile. Not Type. Cancer Discov 2016;6:688. [Crossref] [PubMed]
- Sartore-Bianchi A, Trusolino L, Martino C, et al. Dual-targeted therapy with trastuzumab and lapatinib in HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol 2016;17:738-46. [Crossref] [PubMed]
- Hurwitz H, Hainsworth JD, Swanton C, et al. Targeted therapy for gastrointestinaI (GI) tumors based on molecular profiles : Early results from MyPathway, an open label phase IIa basket study in patients with advanced solid. J Clin Oncol 2016;34:abstr 653.
- Valtorta E, Martino C, Sartore-Bianchi A, et al. Assessment of a HER2 scoring system for colorectal cancer: results from a validation study. Mod Pathol 2015;28:1481-91. [Crossref] [PubMed]
- Pyo JS, Kang G, Park K. Clinicopathological significance and diagnostic accuracy of HER2 immunohistochemistry in colorectal cancer: a meta-analysis. Int J Biol Markers 2016;31:e389-e394. [Crossref] [PubMed]
- Vasan N, Yelensky R, Wang K, et al. A targeted next-generation sequencing assay detects a high frequency of therapeutically targetable alterations in primary and metastatic breast cancers: implications for clinical practice. Oncologist 2014;19:453-8. [Crossref] [PubMed]
- Takegawa N, Yonesaka K, Sakai K, et al. HER2 genomic amplification in circulating tumor DNA from patients with cetuximab-resistant colorectal cancer. Oncotarget 2016;7:3453-60. [PubMed]
- Velho S, Oliveira C, Ferreira A, et al. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur J Cancer 2005;41:1649-54. [Crossref] [PubMed]
- Samuels Y, Wang Z, Bardelli A, et al. Brevia: high frequency of mutations of the PIK3Ca gene in human cancers. Science 2004;304:554. [Crossref] [PubMed]
- De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol 2010;11:753-62. [Crossref] [PubMed]
- Sartore-Bianchi A, Martini M, Molinari F, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res 2009;69:1851-7. [Crossref] [PubMed]
- Mao C, Yang ZY, Hu XF, et al. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. Ann Oncol 2012;23:1518-25. [Crossref] [PubMed]
- Mei ZB, Duan CY, Li CB, et al. Prognostic role of tumor PIK3CAmutation in colorectal cancer: a systematic review and meta-analysis. Ann Oncol 2016;27:1836-48. [Crossref] [PubMed]
- Liao X, Lochhead P, Nishihara R, et al. Aspirin Use, Tumor PIK3CA Mutation, and Colorectal-Cancer Survival. N Engl J Med 2012;367:1596-606. [Crossref] [PubMed]
- Paleari L, Puntoni M, Clavarezza M, et al. PIK3CA Mutation, Aspirin Use after Diagnosis and Survival of Colorectal Cancer. A Systematic Review and Meta-analysis of Epidemiological Studies. Clin Oncol (R Coll Radiol) 2016;28:317-26. [Crossref] [PubMed]
- Ney JT, Froehner S, Roesler A, et al. High-resolution melting analysis as a sensitive prescreening diagnostic tool to detect KRAS, BRAF, PIK3CA, and AKT1 mutations in formalin-fixed, paraffin-embedded tissues. Arch Pathol Lab Med 2012;136:983-92. [Crossref] [PubMed]
- Arsenic R, Treue D, Lehmann A, et al. Comparison of targeted next-generation sequencing and Sanger sequencing for the detection of PIK3CA mutations in breast cancer. BMC Clin Pathol 2015;15:20. [Crossref] [PubMed]
- Li WM, Hu TT, Zhou LL, et al. Highly sensitive detection of the PIK3CA (H1047R) mutation in colorectal cancer using a novel PCR-RFLP method. BMC Cancer 2016;16:454. [Crossref] [PubMed]
- Ang D, O’Gara R, Schilling A, et al. Novel method for PIK3CA mutation analysis: Locked nucleic acid-PCR sequencing. J Mol Diagn 2013;15:312-8. [Crossref] [PubMed]
- Schildgen V, Lüsebrink J, Appel JD, et al. Identification of uncommon PIK3CA mutations in lung cancer by using pyrosequencing. Diagn Mol Pathol 2013;22:22-7. [Crossref] [PubMed]
- Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med 2003;349:247-57. [Crossref] [PubMed]
- Sinicrope FA, Sargent DJ. Molecular Pathways: Microsatellite Instability in Colorectal Cancer: Prognostic, Predictive, and Therapeutic Implications. Clin Cancer Res 2012;18:1506-12. [Crossref] [PubMed]
- Sinicrope FA, Foster NR, Thibodeau SN, et al. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J Natl Cancer Inst 2011;103:863-75. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Snyder A, Makarov V, Merghoub T, et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N Engl J Med 2014;371:2189-99. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Dudley JC, Lin MT, Le DT, et al. Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin Cancer Res 2016;22:813-20. [Crossref] [PubMed]
- Koopman M, Kortman GA, Mekenkamp L, et al. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer 2009;100:266-73. [Crossref] [PubMed]
- Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 2015;21:827. [Crossref] [PubMed]
- Thierry AR, Mouliere F, El Messaoudi S, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 2014;20:430-5. [Crossref] [PubMed]
- Mostert B, Jiang Y, Sieuwerts AM, et al. KRAS and BRAF mutation status in circulating colorectal tumor cells and their correlation with primary and metastatic tumor tissue. Int J Cancer 2013;133:130-41. [Crossref] [PubMed]
- Mohamed Suhaimi NA, Foong YM, Lee DY, et al. Non-invasive sensitive detection of KRAS and BRAF mutation in circulating tumor cells of colorectal cancer patients. Mol Oncol 2015;9:850-60. [Crossref] [PubMed]
- Buim ME, Fanelli MF, Souza VS, et al. Detection of KRAS mutations in circulating tumor cells from patients with metastatic colorectal cancer. Cancer Biol Ther 2015;16:1289-95. [Crossref] [PubMed]
- Lyberopoulou A, Aravantinos G, Efstathopoulos EP, et al. Mutational analysis of circulating tumor cells from colorectal cancer patients and correlation with primary tumor tissue. PLoS One 2015;10:e0123902. [Crossref] [PubMed]
- Spindler KL, Pallisgaard N, Vogelius I, et al. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res 2012;18:1177-85. [Crossref] [PubMed]
- Spindler KL, Pallisgaard N, Appelt AL, et al. Clinical utility of KRAS status in circulating plasma DNA compared to archival tumour tissue from patients with metastatic colorectal cancer treated with anti-epidermal growth factor receptor therapy. Eur J Cancer 2015;51:2678-85. [Crossref] [PubMed]
- De Miglio MR, Mura A, Uras MG, et al. High sensitivity of reverse-hybridization methodology in the detection of KRAS mutations from formalin-fixed paraffin-embedded colorectal cancer samples. Diagn Mol Pathol 2010;19:201-8. [Crossref] [PubMed]
- Fariña Sarasqueta A, Moerland E, De Bruyne H, et al. SNaPshot and StripAssay as valuable alternatives to direct sequencing for KRAS mutation detection in colon cancer routine diagnostics. J Mol Diagn 2011;13:199-205. [Crossref] [PubMed]
- Weckselblatt B, Rudd MK. Human Structural Variation: Mechanisms of Chromosome Rearrangements. Trends Genet 2015;31:587-99. [Crossref] [PubMed]
- Schenkel LC, Kerkhof J, Stuart A, et al. Clinical Next-Generation Sequencing Pipeline Outperforms a Combined Approach Using Sanger Sequencing and Multiplex Ligation-Dependent Probe Ampli fi cation in Targeted Gene Panel Analysis. J Mol Diagn 2016;18:657-67. [Crossref] [PubMed]
- Wang Q, Xia J, Jia P, et al. Application of next generation sequencing to human gene fusion detection: Computational tools, features and perspectives. Brief Bioinform 2013;14:506-19. [Crossref] [PubMed]
- Chmielecki J, Peifer M, Jia P, et al. Targeted next-generation sequencing of DNA regions proximal to a conserved GXGXXG signaling motif enables systematic discovery of tyrosine kinase fusions in cancer. Nucleic Acids Res 2010;38:6985-96. [Crossref] [PubMed]
- Meric-Bernstam F, Brusco L, Shaw K, et al. Feasibility of Large-Scale Genomic Testing to Facilitate Enrollment Onto Genomically Matched Clinical Trials. J Clin Oncol 2015;33:2753-62. [Crossref] [PubMed]
- Chantrill LA, Nagrial AM, Watson C, et al. Precision medicine for advanced pancreas cancer: The individualized molecular pancreatic cancer therapy (IMPaCT) Trial. Clin Cancer Res 2015;21:2029-37. [Crossref] [PubMed]
- Boland GM, Piha-Paul SA, Subbiah V, et al. Clinical next-generation sequencing to identify actionable alterations in a phase I program. Oncotarget 2015;6:20099-110. [Crossref] [PubMed]
- Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998-2006. [Crossref] [PubMed]
- Overman MJ, Morris V, Kee B, et al. Utility of a molecular pre-screening program in advanced colorectal cancer for enrollment on biomarker-selected clinical trials. Ann Oncol 2016;27:1068-74. [Crossref] [PubMed]
- United States National Institute of Health. ClinicalTrials.gov - NCT02465060, NCI-MATCH: Targeted Therapy Directed by Genetic Testing in Treating Patients with Advanced Refractory Solid Tumors or Lymphomas (cited 2016 Aug 1). Available online: http://linkinghub.elsevier.com/retrieve/pii/S009082581530175X


