The anatomy of the TNM for colon cancer
Introduction
The tumor, lymph node, and metastasis (TNM) staging system has undergone seven revisions since first publication of the Cancer Staging Manual in 1977 (1). These revisions were vital in order to address improvements in oncology including advancements in early detection, patient management, treatment, and discovery of new prognostic and predictive factors. These revisions, however, revealed the challenges associated with the development of clinical systems of classification in view of the rapid application of research findings (2). For integration into medical practice, clinical prognostic systems have incorporated a number of approaches, including tree based methods (3,4), nomograms (5,6), and dendrograms (7,8). Dendrograms are constructed as binary tree-like diagrams that reveal the similarity among objects or clusters of objects, usually through pair-wise comparisons. For this publication, we explored the advantages of the dendrogram as a novel method for visualizing the stage groups of colon cancer. To emphasize their utility we compared dendrograms constructed from the TNM with Dukes’ system of staging. The dendrograms may offer an additional method for testing and visualizing new systems for patient stratification.
Methods
Data source
Cases of colon cancer were obtained from the Surveillance, Epidemiology, and End Results (SEER) program of the National Cancer Institute from 2004 through 2008 (9). This allowed for follow-up to 2013. After exclusions, 129,174 cases were available for study. Cases of in-situ carcinomas, “Tis,” were excluded. To insure uniformity in histologic type, all epithelial tumors were selected including invasive tumors arising in polyps. However, for patients to be listed in SEER, formal hospital admission is required. Therefore, tumors found in polyps removed in outpatient settings are not recorded. More than 90% of epithelial tumors were listed as some type of “adenocarcinoma”. Non-epithelial tumors and carcinomas originating in the rectum were not included. All cases were accepted even if colon cancer was not the first primary tumor or if followed by a second primary cancer in another site. Cases diagnosed by autopsy or death certificate were excluded due to limited information. SEER does not report subdivisions of the primary stage groupings, for instance the “m” or “y’ categories. In addition, SEER does not report treatment information. Histologic tumor types were coded according to the International Classification of Disease for Oncology (10). Carcinoma, NOS [8010]; adenocarcinoma, NOS [8140]; signet ring cell carcinoma [8490]; adenocarcinoma in adenomatous polyp [8210]; adenocarcinoma in villous adenoma [8261]; adenocarcinoma in tubulo-villous adenoma [8263]; mucinous carcinoma [8480]; and mucin producing adenocarcinoma [8481].
Dendrograms
To show the relationship between combinations of prognostic factors and survival, a hierarchical clustering algorithm referred to as the “ensemble algorithm of clustering of cancer data” (EACCD) was used (7). The output of the algorithm is a tree-structured dendrogram that organizes and stratifies patients according to disease specific survival only. The algorithm was written in the programming language “R”, an open source code available on the Internet (11). The dendrogram represents a visual relationship among survival rates of patients with different combinations of prognostic factors. Because dendrograms cluster combinations according to survival only, they are able to group different combinations of prognostic factors. Cutting the dendrogram at a specified height along its “dissimilarity” axis generates groups of combinations, where combinations from the same group have a “similar” aggregate survival. The dissimilarity only compares survival rates between groups; it does not address survival itself. Details on the algorithm and the creation of dendrograms have been published (7).
The algorithm generated dendrograms based on prognostic factors and survival rates of individual patients listed in the file “case records’ of the SEER program. A dendrogram, representing a tree-diagram of hierarchical clustering, utilizes (the learnt) dissimilarity to measure the difference between two survival rates associated with two combinations of any number of prognostic factors. The dissimilarity values range from 0.0 to 1.0, with larger values of dissimilarity denoting larger differences in outcome between two patient cohorts.
Computationally, the dendrogram is created iteratively through a series of merging steps from bottom to top. First, each combination of prognostic factors is treated as a distinct cluster. Then, at each successive step, two clusters that have the least dissimilarity are merged into a larger single cluster, a process of pairwise comparison. The resultant dendrogram represents a clustering tree-like structure where each node, now reading from top to bottom, apportions the merged clusters into branches that define unfavorable (relatively aggressive) and favorable (relatively indolent) prognostic cohorts based on survival and biologic patterns (7).
As an additional advantage, the algorithm circumvents the limitation on the number of factors that can be added to the TNM (2), because it assigns prognostic groups only based on survival and not on a predefined rule-based system for extent of disease and survival. It also takes into account censored survival times in contrast to traditional dendrogram generators, which are unable to deal with censoring. The algorithm can accept any type of prognostic factor (e.g., continuous, ordinal, or nominal) including molecular factors and sets no limit on the number of factors or the sequence at which they are entered.
Prognostic factors
A combination of prognostic factors is defined as a group of any number of factors. For example, two colonic cancers, both T1, N1, M0, but with different locations in the colon, represent two combinations. As the number of factors increases, the number of patients in a cohort or reference dataset must also increase to accommodate all combinations in order to achieve statistical significance. SEER seemed a reasonable choice as a reference dataset for demonstration because of its large size, high level of quality control, long history, wide geographic coverage, lifetime follow-up, and unbiased ascertainment.
Survival analysis
The algorithm generates a spectrum of survival rates for every combination of prognostic factors. For example, for colon cancer if we use the standard 4 “T” categories, 3 “N” categories, 2 “M” categories, then 24 combinations (4×3×2) are generated. Adding tumor grade, (four categories) increases the number of combinations to 96. Therefore, 96 survival rates will be calculated, one for each combination. In practice, the number of survival rates is usually less than the number of combinations because some combinations, such as a T1, N0, M1, are rarely found in the colon. Furthermore, combinations that contained fewer than 50 patients were arbitrarily excluded because of the approximation of the chi-square distribution in the algorithm. Any reasonable number, however, can be excluded. Disease specific survival rates were calculated by the Kaplan Meier method (12,13).
Results
Anatomy of a staging system
Using dendrograms, the anatomy of the TNM for colon cancer is shown step by step starting with a single factor. Figure 1 shows a dendrogram for the four “T” categories only, and Figure 2 the 5-year survival rates for each of the categories. As expected, T1 and T2 cancers are more similar to each other than they are to the cancers with transmural invasion or extension to the peritoneal surface. In Figure 2, the survival rates of T1 and T2 are statistically similar according to the log rank test, P<0.05, but the rates of T3 and T4 are statistically different.
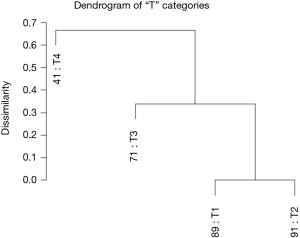
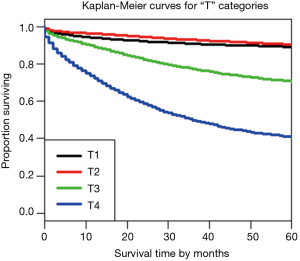
Figure 3 shows the dendrogram for nodal status only and Figure 4 the 5-year survival rates for the three nodal categories. The dendrographic display demonstrates that N1 and N2 are markedly distinct from tumors with N0 involvement.
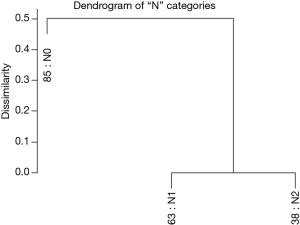
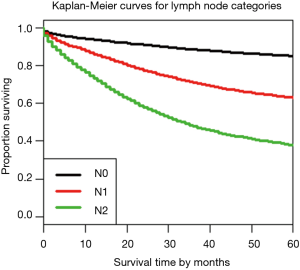
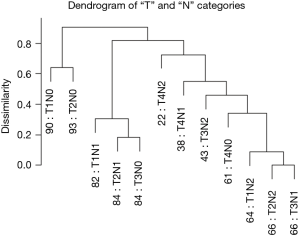
Figure 5 reveals a dendrogram for the T and N categories combined, which has 12 combinations (3×4) and recapitulates Dukes’ classification (14-16). In order to compare with AJCC, we had to convert Dukes’ to the “T, N” definitions. Figure 5 also shows that dendrograms may produce branches that cluster mixed stage subgroups. Though stage I (T1, N0 and T2, N0) co-cluster, it is apparent that some stage II cases (T3, N0) track near early stage III (T1, N1 and T2, N1). This feature seems to indicate that stage II represents a mixed group of potentially aggressive tumors that may benefit from chemotherapy, appropriate for stage III cancers, and a relatively indolent group that may not benefit from chemotherapy. Similarly, stage II (T4, N0) cancers are found within a larger cluster of stage III cancers.
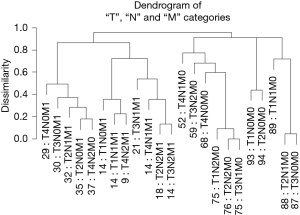
Combining the T, N, and M variables produces a more complex dendrogram (Figure 6) that represents a visualization of the 6th edition AJCC staging system for colon cancer (17). The dendrogram contains 23 combinations instead of the 24 since one combination that did not contain at least 50 patients was excluded. The projected 24 combinations arise from the interaction of 4 T categories, 3 N categories, and 2 M categories (4×3×2) of the TNM. Once again, we have excluded “Tis” and considered all epithelial tumor types and all divisions of the colon except the rectum. Because of pairwise comparisons, the dendrogram pattern in Figure 6 shows that an initial bifurcation results in a more favorable outcome with lower TNM stages on the right side and higher stages with less favorable outcomes on the left branches. No tumors that contain metastatic spread are present on the major right branches. A number of different combinations of T, N, and M are associated with similar survival rates, which have implications for the conduct of clinical trials.
Prognostic groups
By cutting the dendrogram along its dissimilarity axis, prognostic subgroups are generated according to survival rates, which are significantly different from each other.
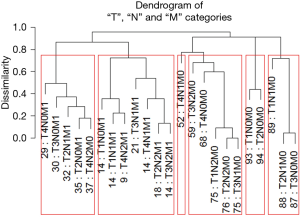
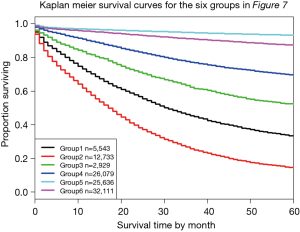
Figure 7 shows Figure 6 arranged into six groups by cutting the dendrogram at a dissimilarity value of approximately 0.75. The 5-year survival rate for each group is shown in Figure 8. The rates in Figure 8 are statistically different without overlap and would fulfill criteria for a modified staging system. However, these groupings contain patients with different combinations of prognostic factors, which results from stratifying only on survival.
Multiple prognostic factors and survival
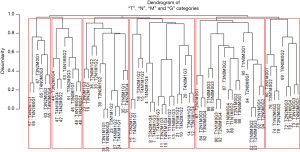
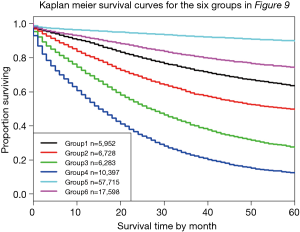
Figure 9 shows a dendrogram generated from 96 combinations (4 T categories, 3 N categories, 2 M categories, and 4 histological grades, which serve as the additional factor integrated into TNM). The dendrogram has been divided into six groups by cutting along the dissimilarity axis around 0.95. The survival rate for each group is presented in Figure 10. Any two of the six survival rates are statistically different (P<0.05). There is no overlap or crossover in the rates, even though each group contains different combinations of prognostic factors. The meaningful combinations are partitioned so that those with similar survival rates are grouped together. Any number of groups can be generated depending on the level at which the dendrogram is cut along its dissimilarity axis. In principle, each prognostic group could serve as a stage group although the prognostic groups would reflect the effect of multiple prognostic factors along with extent of disease. Because of pairwise comparisons, a number of different combinations of prognostic factors have similar outcomes (Figure 9), which we have consistently observed and which has implications for the conduct of clinical trials. For instance, outcome similarities are seen for T3, N1, M1, G1 and T4, N2, M0, G3.
Discussion
Our research demonstrates that an algorithmic approach, based on cluster analysis, is able to utilize a national cancer database to visualize the interaction of prognostic factors. Cluster analysis has been applied in many pursuits, such as image processing, biology, medicine, and others (7). It has the potential for multiple applications in cancer including patient stratification, validation of new staging systems, and application to cancers that do not have TNM factors such as lymphomas.
We have described how the EACCD, an unsupervised learning algorithm, can create prognostic groups based on combinations of any number and type of factors including molecular factors (7,8). The process involves (I) generating dendrograms from all combinations of prognostic factors; and (II) sorting the dendrograms into prognostically relevant patterns according to survival. As a result, it becomes possible to amplify TNM or similar staging systems with additional factors without changing current stage definitions. We have hypothesized that an algorithmic approach is able to amplify cancer patient staging and improve estimations of outcome, while the computer model should provide insights into staging beyond that achieved from anatomic based systems.
Dendrograms have the potential to validate different systems of staging. Compared to the AJCC, the Dukes’ system, for instance, is unbalanced since only 2 factors, (T1, N0 and T2, N0) are on the left in the first pairwise comparison (Figure 5). T1, N0 and T2, N0 correspond to Dukes’ “A” classification. In contrast, including distant metastasis, such as the AJCC, provides a balance for the system as shown in Figure 6 for the TNM. Cases that are T4 or M1 are located on the dendrogram’s left and cases with M0 on the right. Most importantly, note the differences in outcome for similar TN combinations. For instance, for T4, N1, Dukes’ predicts a survival of 38% while the AJCC’s T4, N1, M0 predicts a survival of 52%. By not including the “M” categories, in this case M1, predictions made by Dukes’ actually include both “M0” and “M1”. By including either “M0” or “M1” only as in the TNM, the non-included “M”, rather “M0” or “M1”, is excluded and its effect on survival not considered. All physicians, however, know the consequences of distant metastasis. Nonetheless, the purpose of Dukes’ and the AJCC’s system are essentially identical. They assess disease severity in order to plan appropriate therapy.
The progressive development of the TNM is seen in Figures 1-8. Given a “T1” tumor, the dendrograms inform us about the effect of progressive nodal involvement on the “T” category. Moreover, with dendrograms, the effect of one prognostic factor on survival can be visually tested by varying the factor and fixing remaining factors at constant categories. Assuming successful validation, an algorithmic approach is expected to have a significant application for personalized medicine because disease specific outcomes for individual combinations of prognostic factors can be given so that outcome predictions for new cancer patients can be based on previous patients who have had similar combinations of prognostic factors.
Since the algorithm clusters combinations of prognostic factors based on survival rates only, combinations composed of different factors could show somewhat similar survival. For example, T3, N1, M1, G1 (31% 5-year survival) has the same outcome as T4, N2, M0, G3 (33% 5-year survival) as seen in Figure 9. When comparing results of clinical studies, the prognostic factors should be compared along with survival. As we indicated previously, different combinations with similar survival have implications for the design and interpretation of clinical trials (18). Cohorts designated by tumor stage alone are likely to contain multiple different prognostic combinations that affect survival. Different distributions of these factors among cohorts of the same stage group may increase or decrease the effect of treatment (18).
Finally, we should stress that by stratifying only on survival, unlimited additional prognostic factors can be integrated into the TNM or in other systems of classification. We integrated the histological grade as an example of an additional factor (Figure 9). However, we have been able to integrate multiple factors (7). Stratifying patients according to multiple and dissimilar prognostic factors with similar outcome should provide more homogeneous populations for clinical trials. More importantly, a true treatment effect will likely be detected with homogeneous populations than with heterogeneous populations. Based on our previous research, we have concluded that survival is relative and depends on the prognostic factors selected (18).
Acknowledgements
The authors would like to thank Ms. Linh Nguyen for her wonderful assistance that led to completion of this manuscript.
Funding: D Chen, MT Hueman and DE Henson were partially supported by the grant “Using Dendrograms to Create Prognostic Systems for Cancer” (No. 307170) sponsored by the John P. Murtha Cancer Center Research Program at the Walter Reed National Military Medical Center and the Uniformed Services University of the Health Sciences. AM Schwartz was supported by the grant “Prognostic Markers in Early Stage Lung Cancer: Computer Algorithms and Bayesian Regression” sponsored by the Dr. Cyrus and Myrtle Katzen Cancer Research Grant Award at The George Washington University.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Disclaimer: The views expressed by the authors do not necessarily reflect the official views of the Uniformed Services University of the Health Sciences, the Department of Defense, or the U.S. Government.
References
- Edge SB, Byrd DR, Compton CC, et al. editors. AJCC Cancer Staging Manual. 7th ed. New York, NY: Springer-Verlag, 2009.
- Burke HB, Henson DE. The American Joint Committee on Cancer. Criteria for prognostic factors and for an enhanced prognostic system. Cancer 1993;72:3131-5. [Crossref] [PubMed]
- Gimotty PA, Guerry D, Ming ME, et al. Thin primary cutaneous malignant melanoma: a prognostic tree for 10-year metastasis is more accurate than American Joint Committee on Cancer staging. J Clin Oncol 2004;22:3668-76. [Crossref] [PubMed]
- Radespiel-Tröger M, Hohenberger W, Reingruber B. Improved prediction of recurrence after curative resection of colon carcinoma using tree-based risk stratification. Cancer 2004;100:958-67. [Crossref] [PubMed]
- Kattan MW, Reuter V, Motzer RJ, et al. A postoperative prognostic nomogram for renal cell carcinoma. J Urol 2001;166:63-7. [Crossref] [PubMed]
- Liang W, Zhang L, Jiang G, et al. Development and validation of a nomogram for predicting survival in patients with resected non-small-cell lung cancer. J Clin Oncol 2015;33:861-9. [Crossref] [PubMed]
- Chen D, Hueman MT, Henson DE, et al. An algorithm for expanding the TNM staging system. Future Oncol 2016;12:1015-24. [Crossref] [PubMed]
- Chen D, Xing K, Henson D, et al. Developing prognostic systems of cancer patients by ensemble clustering. J Biomed Biotechnol 2009;2009:632786.
- National Cancer Institutes. Surveillance, Epidemiology, and End Results Program. SEER*Stat Software. 2016. Availale online: www.seer.cancer.gov/seerstat
- World Health Organization. International Classification of Diseases for Oncology, 3rd Edition (ICD-O-3). Available online: http://www.who.int/classifications/icd/adaptations/oncology/en/
- The R Project for Statistical Computing. Available onlne: http://www.r-project.org
- Kaplan EL, Meier P. Nonparametric Estimation from Incomplete Observations. J Am Stat Assoc 1958;53:457-81. [Crossref]
- Klein JP, Moeschberger ML. Survival Analysis: Techniques for Censored and Truncated Data. 2nd edition. New York: Springer-Verlag New York, 2003.
- Dukes CE. The classification of cancer of the rectum. J Pathol Bacteriol 1932;35:323-32. [Crossref]
- Dukes C. The spread of cancer of the rectum. Br J Surg 1929-30;17:643-8.
- Gabriel WB, Dukes C, Bussey HJ. Lymphatic spread in cancer of the rectum. Br J Surg 1935;23:395-413. [Crossref]
- Greene FL, Page DL, Fleming ID, et al. editors. AJCC Cancer Staging Manual. 6th edition. New York, NY: Springer-Verlag New York, 2002.
- Henson DE, Schwartz AM, Chen D, et al. The clinical implications of integrating additional prognostic factors into the TNM. J Surg Oncol 2014;109:391-4. [Crossref] [PubMed]


