Predictive and prognostic biomarkers in personalized gastrointestinal cancer treatment
Introduction
Gastrointestinal cancers are among the leading causes of cancer morbidity and mortality worldwide. Although many chemotherapeutic agents and combinations are available, the overall prognosis of patients with advanced gastrointestinal cancer remains poor.
Distinct molecular pathways have been identified in each tumor type. This deeper knowledge of molecular pathology offers the opportunity to develop novel targeted personalized therapies. However when tested in an unselected population, many targeted drugs show limited efficacy, rendering patient selection and biomarkers a critical need to optimally employ these treatments. As genetics-based oncology moves forward, medical decisions will become increasingly linked to genomic and proteomic tumoral features.
Biomarkers are biological molecules found in tissues, blood, and other body fluids. They are signs representing a normal or abnormal process, or a condition or disease. Prognostic biomarkers are associated with the clinical outcome and are used to identify patients with a more aggressive disease course. Predictive biomarkers are measures of the likelihood of response or lack of response of a particular therapy, and allow identification of patients most likely to benefit from a given treatment, thus sparing other patients from toxicities of ineffective therapies.
In gastrointestinal cancers, biomarkers have contributed to the approval of several new targeted therapies. In colorectal cancer (CRC), various epidermal growth factor receptor (EGFR) targeted therapies have been approved for patients selected in terms of their rat sarcoma-2 virus (RAS) status. Other drug developments have been made based on particular molecular mechanisms; hence, with the knowledge of a particular molecular alteration, population has been selected. For example, development of anti-human epidermal growth factor receptor 2 (HER2) treatments in gastric cancer has been done in the selected HER2 positive population.
This review provides insights into the current status of the biomarkers used in gastrointestinal cancers—CRC, gastric and pancreatic—and describes the most promising biomarkers based on the molecular characteristics of each tumor type.
CRC
CRC is one of the most prevalent cancers worldwide, representing approximately 10% of all cancer diagnoses (1). It is the second leading cause of cancer-related deaths worldwide. However, progress in treatment modalities and advances in molecular biomarkers allowing personalized treatments have together contributed to improvements in overall survival (OS) of patients with metastatic CRC (mCRC).
In recent years, CRC has been extensively characterized from a molecular perspective. Several molecular hallmarks have been identified and subgroups with prognostic and predictive implications have been proposed. Analyses of the molecular profile have revealed CRC to be a heterogeneous disease with several genes commonly mutated, including Kirsten rat sarcoma-2 virus (KRAS), neuroblastoma-RAS (NRAS), BRAF and phosphatidylinositol-4,5-bisphosphonate 3-kinase, catalytic subunit alpha polypeptide gene (PIK3CA). CRC has classically been divided into two subgroups with distinct prognostic and treatment connotations; stable tumors and tumors with microsatellite instability (MSI). These genetic alterations and classifications are used to guide a personalized treatment approach and predict disease outcomes.
RAS in CRC
To date, the only accepted negative predictive biomarker in the treatment of mCRC is the RAS proto-oncogene mutation. KRAS encodes a small guanosine triphosphate (GTP) binding protein that acts as a self-inactivating signal transducer by cycling from guanosine diphosphate (GDP)- to GTP-bound states (2). RAS mutations can result in permanent activation of signaling pathways that control cell proliferation, differentiation, adhesion, apoptosis, and migration. KRAS is a member of the RAS/RAF/mitogen-activated protein kinase (MAPK) and PI3K/AKT (protein kinase B) signaling pathways, which are activated by EGFR (3). Such mutations are found in approximately 30% to 50% of all CRC tumors (4). Three RAS genes have been linked to cancer: HRAS, KRAS and NRAS (2). Mutations in KRAS and NRAS are found in approximately 50% of advanced CRCs, with the majority of mutations involving KRAS exon 2 (90%) (5).
Initial clinical trials assessing the efficacy of agents targeting EGFR in CRC were performed in unselected population (6,7). Subsequently, the presence of mutations in KRAS codons 12 and 13 evaluated in clinical trials with panitumumab and cetuximab monotherapy were found to be predictive of a lack of response (8,9) (Table 1). A phase 3 clinical trial evaluating panitumumab plus best supportive care versus best supportive care alone in patients with chemotherapy-refractory mCRC was performed. The primary end point was progression-free survival (PFS) and secondary endpoints included objective response, OS and safety (7). Patients in the best supportive care arm who progressed could subsequently receive panitumumab. Panitumumab was found to significantly prolong PFS, with a hazard ratio (HR) of 0.54 [95% confidence interval (CI), 0.44–0.66, P<0.0001]. The predictive role of KRAS was evaluated, with median PFS improved by the addition of panitumumab to best supportive care compared with best supportive care alone in patients with wild-type KRAS exon 2 (12.3 vs. 7.3 weeks, respectively; HR 0.45; 95% CI, 0.34–0.59). In patients with mutant KRAS, PFS was not significantly improved with panitumumab (8).

Full table
Similar outcomes were reported with cetuximab in a randomized trial compared with best supportive care in chemorefractory mCRC patients. The primary endpoint was OS, which was significantly improved with a HR of 0.77 (95% CI, 0.54–0.92, P=0.005) (11). KRAS mutational status was evaluated in 394 of 572 patients (69%) showing that the effectiveness of cetuximab was significantly associated with KRAS mutation status. In patients with wild-type KRAS tumors, treatment with cetuximab significantly improved OS as compared with supportive care alone (median, 9.5 vs. 4.8 months; HR 0.55; 95% CI, 0.41–0.74; P<0.001) and PFS (median, 3.7 vs. 1.9 months; HR 0.40; 95% CI, 0.30–0.54; P<0.001). Among patients with mutated KRAS tumors, there was no significant difference between those who were treated with cetuximab and those who received supportive care alone with respect to OS (HR 0.98, P=0.89) or PFS (HR 0.99, P=0.96). Among patients receiving best supportive care alone, the mutational status of the KRAS gene was not significantly associated with OS (HR 1.01, P=0.97) (9).
Subsequent clinical trials assessing cetuximab and panitumumab in combination with chemotherapy in the first-line setting also tested KRAS status, confirming the lack of benefit in patients harboring KRAS mutations (12,13). Based on these results, anti-EGFR therapy was restricted to the KRAS wild-type population (14).
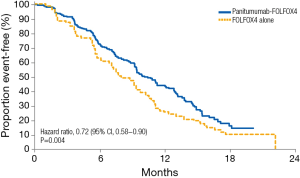
Analyses evaluating the role of mutations beyond KRAS exon 2 in the context of anti-EGFR agents have also been performed (15). Early retrospective RAS analyses led to the hypothesis that these mutations have additional predictive value in terms of clinical outcomes (16,17). Results from the extended RAS analyses of randomized phase 3 studies in mCRC patients treated with chemotherapy and anti-EGFR antibodies provided evidence supporting appropriate patient selection on the basis of RAS mutation status. The PRIME clinical trial evaluated the efficacy of panitumumab in combination with oxaliplatin, fluorouracil and leucovorin (FOLFOX4) in KRAS exon 2 wild-type patients. In a prospective-retrospective analysis the treatment effect of panitumumab was evaluated according to RAS mutational status, including KRAS and NRAS exons 2, 3, and 4. In patients without RAS mutations, panitumumab plus FOLFOX4 was associated with significant improvements vs. FOLFOX4 alone in PFS (10.1 vs. 7.9 months; HR 0.72; 95% CI, 0.58–0.90; P=0.004) and OS (26.0 vs. 20.2 months; HR 0.78; 95% CI, 0.62–0.99; P=0.04) (Figure 1) (5). In patients with wild-type KRAS exon 2 tumors and other RAS mutations, panitumumab plus FOLFOX4 did not improve PFS (HR 1.28; 95% CI, 0.79–2.07; P=0.33) or OS (HR 1.29; 95% CI, 0.79–2.10; P=0.31). These data represented the first demonstration in a phase 3 study of the value of extended RAS analysis for anti-EGFR therapy.
KRAS status has also been evaluated as a prognostic biomarker. A recent analysis of the MAVERICC trial evaluated the prognostic role of KRAS (18). This phase 2 randomized clinical trial compared the combination of FOLFOX plus bevacizumab vs. FOLFIRI plus bevacizumab, with KRAS found to be a prognostic biomarker. In wild-type patients, median OS was 26.1 months in the FOLFOX arm and 36.7 months in the FOLFIRI arm, compared to 22.5 months and 26.9 months in the mutant KRAS group, respectively. This study contributes to the growing evidence of the prognostic role of KRAS mutation.
In relation to non-invasive biomarkers, several studies have investigated the use of circulating tumor DNA to analyze the mutational status, most studying KRAS. The largest evaluation of circulating tumor DNA was in the context of the phase 3 CORRECT trial assessing the activity of regorafenib compared to placebo (19). Mutational analysis was done with BEAMing, a technique based on emulsion polymerase chain reaction (PCR), to identify KRAS, PIK3CA and BRAF in DNA from the plasma of 503 patients. KRAS mutations were identified in 349 (69%) patients, PIK3CA mutations in 84% (17%) and BRAF mutations in 17 (3%) patients. Among patients who received anti-EGFR therapy and with KRAS wild-type archived tumor tissue, 48% were identified as having KRAS mutations with BEAMing analysis. Clinical benefit was seen with regorafenib irrespective of KRAS mutation status. The findings from this study support BEAMing analysis of circulating DNA as a viable approach to obtain tumor-associated genotypic information in a non-invasive manner. This is of particular interest given that the mutational profile in the tumor sample obtained at diagnosis may not represent the actual genotype after multiple lines of treatment. Furthermore, those analyses also offer the possibility of detecting mutations that confer drug resistance.
To summarize, RAS status is a negative predictive biomarker of anti-EGFR treatment efficacy and also plays a prognostic role in mCRC. RAS mutations are now widely routinely tested to determine the most appropriate treatment in mCRC patients, with an emerging trend to evaluate the utility of circulating tumor DNA in the future.
BRAF in CRC
BRAF is the principal effector of RAS in the signaling cascade. Activating BRAF mutations are detected in about 10% to 15% of all mCRC cases (20). The most frequently reported BRAF tumor mutation is a valine-to-glutamic acid amino acid substitution (V600E) that leads to constitutive activation of the MAPK signaling cascade. BRAF mutations are an early event in colorectal tumorigenesis and are associated with the transformation of epithelia to traditional serrated adenomas and sessile serrated adenomas (21,22). BRAF mutations are associated with MSI; in sporadic CRC, BRAF mutations are found in about 40% to 60% of MSI tumors, compared to in only approximately 5% to 10% of microsatellite stable tumors (23). BRAF and KRAS mutations tend to be mutually exclusive (24).
The prognostic role of BRAF has been explored in several retrospective studies and clinical trials. The BRAF V600E mutation has been associated with a poor prognosis in mCRC. In a study published by Samowitz et al., a cohort of 911 patients with stage I to IV CRC, the 5-year OS of patients with BRAF-mutant CRC was significantly lower compared to wild-type tumors (47.5% vs. 60.7%, P<0.01) (25).
The predictive role of BRAF mutations in terms of EGFR targeted agents is not well established. Several studies have suggested that the BRAF mutation confers poor outcomes to anti-EGFR therapies. In studies designed to evaluate the efficacy of anti-EGFR antibodies, BRAF-mutated patients did not show a benefit with anti-EGFR treatment. Nevertheless, some studies have failed to show a negative relationship between the BRAF mutation and response. However the small sample size in these studies makes it difficult to draw conclusions (10,17,26).
To design effective strategies to improve BRAF-mutant CRC outcomes, combinations of BRAF inhibitors with other targeted agents have been studied with encouraging preliminary efficacy. In a phase 1–2 trial of dabrafenib (an oral selective inhibitor of BRAF kinase) and trametinib (an elective inhibitor of mitogen-activated protein kinase 1 (MEK1) and MEK2) in BRAF V600E mutated CRC, 12% of patients achieved a partial response or better, including one patient who achieved a durable complete response (27). In another phase 1–2 trial, mCRC patients were treated with dabrafenib and panitumumab or dabrafenib, panitumumab and trametinib (28). The dabrafenib and panitumumab combination gave a 10% response rate and the triple combination gave a 26% rate. Another phase 1b/2 trial assessed the combination of encorafenib (a new-generation BRAF inhibitor) with cetuximab with or without the α-specific PI3K inhibitor alpelisib in CRC patients (29). PFS was 5.4 months with the triplet and 4.2 months with the doublet. The dual arm showed a 22% overall response rate (ORR) and the triple arm a 27%. Taking all these studies into consideration, BRAF mutation is becoming a biomarker for response to combination regimens of BRAF inhibitors and targeted agents.
MSI
Microsatellites are short repetitive DNA sequences located at multiple sites of the genome. The number of repeats contained in any one particular microsatellite is the same in every cell. MSI occurs when some cells display one or two alleles with different numbers of repeats (30). MSI is present in 15–20% of CRC (31). It can be due to germline loss-of-function in the mismatch repair (MMR) genes MLH1, MSH2, PMS2 or MSH6, or due to hypermethylation in the promoter of the MLH1 gene, i.e., it can have a genetic or sporadic origin. Lynch syndrome is an autosomal dominant disorder due to germline loss-of-function mutations in the MMR genes, associated with a predisposition to multiple types of malignancies including cancers of the colon, rectum, endometrium, stomach and small bowel. Approximately 3% of all CRC are Lynch syndrome related. Detection of MSI or MMR is not only used as prescreening for Lynch syndrome, but also to predict prognosis and response to treatment. Several studies including two separate pooled analyses have evaluated the impact of MSI in terms of prognosis (31,32). MSI is associated with a favorable outcome, especially in patients with stage II or III colon cancer. MSI is also suggested to be a negative predictive biomarker for adjuvant 5-fluorouracil-based therapy (33,34).
Data addressing the relationship between MSI and response to immunotherapy are emerging. The high mutational load in MSI tumors creates many tumor-specific neo-antigens (35). Furthermore, MSI tumors have a higher level of tumor infiltrating lymphocytes (36). MSI tumors highly upregulate the expression of multiple immune checkpoints, including programmed death-1 (PD-1), PD ligand 1 (PD-L1), cytotoxic T lymphocyte-associated antigen 4 (CTLA-4), lymphocyte activation gene 3 (LAG-3) and indoleamine 2,3-dioxygenase (IDO) (35).
Although anti-PD-1 and anti-PD-L1 antibodies have demonstrated efficacy in melanoma, renal cancer and lung cancer, CRC has been considered a poor responder. This may however have been due to use of unselected CRC populations in the relevant clinical trials. Since MSI have high expression of immunocheckpoints, anti-PD1 and anti-PD-L1 antibodies might have better outcomes in this subset of patients. Based on this hypothesis, a phase 2 clinical trial was performed in 41 patients with metastatic tumors (37). Pembrolizumab (an inhibitor of the PD-1 immune checkpoint) was administered to patients with MMR deficient CRC, MMR proficient CRC and patients with non-CRC MMR deficient cancers. The immune-related objective response rate was 40% in MMR deficient and 0% in MMR proficient CRC. Median PFS and OS were not reached in the cohort with MMR deficient CRC and were 2.2 and 5.0 months respectively in the cohort with MMR proficient CRC (HR for disease progression or death, 0.1, P<0.001). MSI CRC patients are thus considered good candidates for immunotherapy approaches and clinical trials have been initiated in this patient subset. The outcome should help clarify optimal treatment approaches in MSI patients.
HER2 in CRC
HER2 protein overexpression and HER2 gene amplification are relatively rare in CRC. Evaluation of the predictive value of HER2 status in relation to response to anti-EGFR therapies has given contradictory results. While some studies have shown that HER2 gene amplification is related to resistance to anti-EGFR therapy, others have suggested HER2 status as a marker of benefit from anti-EGFR targeted therapies (38,39).
Recently, a phase 2 trial was performed to assess the efficacy of trastuzumab (a humanized monoclonal antibody that binds HER2) and lapatinib (a dual tyrosine kinase inhibitor which blocks the HER2 and EGFR pathways) in patients with HER2-positive, KRAS exon 2 wild-type mCRC after failure with standard therapies (40). Patients received trastuzumab as a 4 mg/kg loading dose followed by 2 mg/kg once per week and oral lapatinib at 1,000 mg/day until disease progression. Among 914 screened patients with KRAS exon 2 wild-type mCRC, 5% had HER2 positive tumors. Among the 27 HER2 positive patients treated, the ORR was 30% and one patient had a complete response. This study shows the potential predictive value of HER2 in response to anti-HER2 treatment as well as to assess the negative predictive value for anti-EGFR treatment, but confirmatory trials are lacking.
CRC subtyping
Gene expression subtyping is widely accepted as a relevant source of disease stratification. In CRC there are several classifications with discrepant results. Recently, a consortium formed to assess the subtype pattern evaluated the CRC subtyping systems and identified four consensus molecular subtypes (CMS) (41). CMS1, MSI immune, which represents 14% of cases, is characterized by hypermutated tumors, microsatellite unstable and with strong immune activation. CMS2, the canonical subtype, accounting for 37% of cases, are epithelial tumors, chromosomally unstable with marked WNT and MYC signaling activation. CSM3, a metabolic subtype, representing 13% of cases, are epithelial tumors with clear metabolic dysregulation. Finally, CMS4, the mesenchymal subtype, accounting for 23% of cases, has prominent transforming growth factor β activation, stromal invasion and angiogenesis. Samples with mixed features represent 13% of cases and are not classified in any of the four groups. This is a robust classification system that will play a role in prognostic and prediction of drug response in CRC, but still needs to be validated.
Gastric cancer
Gastric cancer is the fourth commonest cause of cancer-related death worldwide (42). Despite recent advances, the prognosis of gastric cancer remains poor, with reported 5-year survival ranging from 20% to 30% (43). Several molecular pathways involved in gastric cancer have been identified, and a molecular classification dividing gastric cancer into four subtypes has been proposed. Tumors positive for the Epstein-Barr virus more frequently have PIK3CA mutations, DNA hypermethylation, and amplification of JAK2, PD-L1 and PD-L2. A second subset, MSI tumors, has elevated mutation rates. The third group, genomically stable tumors, are enriched for RHOA mutations, and finally, tumors with chromosomal instability show aneuploidy and focal amplification of receptor tyrosine kinases (44). This tumor classification provides a deeper knowledge of gastric cancer and an option for patient selection for targeted therapies.
Her2 in gastric cancer
The unique current validated biomarker of response to treatment in gastric cancer is HER2. HER2 belongs to the EGFR family, and is a transmembrane protein with tyrosine kinase activity at its intracellular domain (45). ERBB2 amplification or HER2 overexpression has been reported in 10% to 27% of tumors, and is more frequent in proximal gastroesophageal junction carcinomas and intestinal-type gastric cancer (46).
The value of HER2 as a prognostic factor is a matter of controversy. Some studies have indicated that ERBB2 amplification is associated with poor prognosis and aggressive disease (47), while other reports show no difference in prognosis and do not establish HER2 as an independent prognostic factor (48).
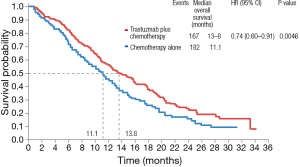
Nevertheless, HER2 is a predictive biomarker of response to trastuzumab. The Trastuzumab for Gastric Cancer trial (ToGA), randomized 584 patients with tumors overexpressing HER2 to receive a fluoropyrimidine (5-FU or capecitabine) plus cisplatin with or without trastuzumab (49). OS was 13.1 months in patients treated with trastuzumab and 11.1 months in patients treated with chemotherapy alone (HR 0.74; 95% CI, 0.60–0.91; P=0.0046) (Figure 2). Following this trial, trastuzumab became the standard of care in patients with tumors overexpressing HER2 and it remains the only validated predictive biomarker for response to targeted therapy in gastric cancer.
Biomarkers of immunotherapy response in gastric cancer
Immune deregulation has been associated with some gastric cancer subtypes, notably those associated with viral infections or with a high mutational rate. As mentioned above the cancer genome atlas (TCGA) proposed a classification of gastric cancer into four subgroups and the immune response to the tumor is likely to play an important role in the Epstein–Barr virus and MSI subgroups (50).
Immune therapies have already been tested in gastric cancer. Safety and activity of pembrolizumab was evaluated in the phase 1 clinical trial KEYNOTE-012. This cohort of 39 gastric cancer patients were positive for PD-L1 (defined as PD-L1 staining in stroma or ≥1% of tumor cells). The overall response rate was 22% by central review. In this study, PD-L1 expression levels were hypothesized to be an effective biomarker (51). Response was correlated with four pre-specified immune-related gene expression signatures, “interferon-gamma”, “TCR-signaling”, “expanded-immune”, and “de novo” signatures (52). These gene expression signatures may be used for the selection of patients more likely to respond to immunotherapy. In addition, two phase 1 clinical trials with durvalumab and atezolizumab (both anti-PD-L1 antibodies) evaluated these agents, including 16 patients and 1 patient with gastric cancer, respectively. Both trials showed a positive correlation between PD-L1 expression and favorable response (50).
In conclusion, although there are no validated biomarkers to select patients who may benefit from immunotherapy, specific gastric cancer subtypes and PD-L1 expression levels are promising biomarkers that need further investigation.
Pancreatic cancer
Pancreatic adenocarcinoma is the fourth leading cause of cancer-related deaths in the US and the Western world (53). Approximately 80% of patients present with advanced disease at diagnosis. This carcinoma has a very poor prognosis with a 5-year survival rate less than 5%.
To better define potential targetable alterations in pancreatic cancer, tumors have been analyzed and its genome has been sequenced. In a study by Biankin and colleagues, exome sequencing was performed to define genomic aberrations in a cohort of stage I and II sporadic pancreatic ductal adenocarcinomas. Sixteen significantly mutated genes were found: KRAS, TP53, CDKN2A, SMAD4, MLL3, TGFBR2, ARID1A and SF3B1, and uncover novel mutated genes including additional genes involved in chromatin modification (EPC1 and ARID2), DNA damage repair (ATM) and other mechanisms (ZIM2, MAP2K4, NALCN, SLC16A4 and MAGEA6) (54). Waddel and colleagues performed whole-genome sequencing and copy number variation analysis in order to classify pancreatic cancer. They identified four subtypes with potential clinical utility: stable, locally rearranged, scattered and unstable. Various mutations of drugable oncogenes such as ERBB2, MET, FGFR1, CDK6, and PIK3CA were found at low individual patient prevalence (55). In terms of biomarkers, although there are no validated biomarkers for patient selection, some molecular alterations with some clinical relevance exist.
BRCA and pancreatic cancer
Familial clustering is found in approximately 10% of pancreatic cancers, suggesting an inherited cancer syndrome (56). In a subset of these families, germline mutations in BRCA1 or BRCA2 genes are present, concurrently conferring a substantially higher risk of breast and ovarian cancer. The relative risk of pancreatic cancer for BRCA1 and BRCA2 mutation carriers has been estimated to be between 2.3 and 7.0 (57). In patients with a familial history of pancreatic cancer, prevalence of the BRCA mutation can be up to 17% (58). In addition, the prevalence of BRCA1 and BRCA2 founder mutations in two retrospective series of Ashkenazi Jewish patients undergoing surgery for pancreatic cancer was found to be 5.5% and 21%, respectively (59,60). Furthermore, in unselected patients, the prevalence of BRCA mutations has been reported to be from 5% to 7% (61,62).
In a study published by Waddell and colleagues (55), whole-genome sequencing and copy number variation analysis of 100 pancreatic ductal adenocarcinomas was performed. Pancreatic cancer was classified into four subtypes based on structural variation profiles and the implicated molecular mechanisms. The unstable subtype was composed of tumors with a large number of structural variation events. This scale of genomic instability suggested defects in DNA maintenance, which potentially defines sensitivity to DNA-damaging agents. The relationship between the unstable subtype, mutations in BRCA pathway genes and a recently described mutational signature associated with deleterious mutations in BRCA1 or BRCA2 in breast, ovarian and pancreatic cancer was analyzed (63). Overlapping deleterious mutations in BRCA1, BRCA2 and PALB2 with unstable genomes and the BRCA mutational signature showed that mutations in these genes were associated with the top quintile of the BRCA mutational signature, and the majority also exhibited unstable genomes. Data from the treatment of patients of this prospective observational cohort study were collected and complemented through therapeutic testing of patient-derived xenografts (PDXs). Overall, eight patients received a platinum-based therapy and seven PDXs were treated with gemcitabine and cisplatin. Of five patients with unstable genomes and/or a high BRCA mutational signature burden, two had exceptional responses and two had robust partial responses. Three patients who did not have any of these characteristics did not respond. These observations were supported by PDX studies, two of three PDX from unstable patients or with high BRCA mutational signature burden also responded to cisplatin. There were no responses in the four PDXs in the other genotype. To sum up, this study suggests that mutations in the BRCA pathway have potential implications for therapeutic selection of pancreatic cancer. These data define a biomarker hypothesis that needs to be tested in a clinical trial.
In the clinical setting, a retrospective analysis of 71 pancreatic cancer patients harboring BRCA mutations investigated the impact of germline BRCA mutations on the natural history and therapeutic outcome with platinum agents (64). A slightly more favorable outcome was observed in the setting of BRCA 1/2 mutation carrier in pancreatic cancer patients. Furthermore, it was also suggested that BRCA-mutated pancreatic cancer patients may benefit from treatment with platinum-based agents.
Poly (ADP-ribose) polymerase (PARP) inhibitors have been studied as potential cancer therapeutics in breast and ovarian cancers. Tumors with an apparent defect in homologous DNA repair seem to be susceptible to PARP inhibitor therapy. This includes tumors with germline or somatic mutations in BRCA1 and BRCA2. In a phase 2 trial, olaparib monotherapy was given to patients with advanced cancer harboring the germline BRCA 1/2 mutation (65). A total of 317 patients were enrolled, 23 of whom had pancreatic adenocarcinoma. Patients with pancreatic cancer received an average of two prior lines of chemotherapy for metastatic disease. The tumor response rate among patients with pancreatic cancer was 21.7% and four patients achieved a complete response. The presence of responses in this heavily pretreated population is of particular interest. This study suggests that patients with pancreatic cancer with mutation in BRCA genes will respond to PARP inhibitors, making the BRCA mutation a potential predictive biomarker of response to PARP inhibitors.
There are currently several trials assessing the effectiveness of PARP inhibitors in patients with pancreatic cancer and BRCA 1/2 mutations. Among them, an ongoing phase 3 trial is evaluating maintenance treatment with olaparib versus placebo in patients who have benefit from a platinum-based therapy (NCT02184195).
Taking all of these data into consideration, although the absence of randomized trials should be noted, BRCA is a potential biomarker of response to platinum-based therapy and PARP inhibitors.
Human equilibrative nucleoside transporter 1 (hENT1) expression in pancreatic cancer
Gemcitabine is one of the most widely used drugs in the treatment of pancreatic cancer. It is hydrophilic and passive diffusion through hydrophobic cellular membranes is thus slow. Permeation through the membranes requires specialized membrane transporters, and hENT1 is the major transporter for gemcitabine (66). Several studies have evaluated the role of hENT1 as a predictive biomarker of response to gemcitabine. The first was a retrospective study analyzing tumor tissue from 21 patients with gemcitabine-treated pancreatic cancer (67). Patients in whom all adenocarcinoma cells had detectable hENT1 had significantly longer median survival from gemcitabine initiation than those without hENT1 in a proportion of adenocarcinoma cells (median survival 13 vs. 4 months, P=0.001). Subsequent studies have provided some evidence for this marker, but were mostly retrospective and non-randomized series with mixed populations (68-70).
Only two randomized trials, with contradictory results, have evaluated the role of hENT1 in the adjuvant setting. One of them, ESPAC3, evaluated 380 patients treated with gemcitabine versus 5-fluorouracil versus observation after resection of pancreatic tumor (71). Median OS for gemcitabine-treated patients was 23.4 months. For patients with low hENT1 expression, median survival was 17.1 (95% CI, 14.3–23.8) months and 26.2 (95% CI, 21.2–31.4) months for those with high hENT1 expression, P=0.002. The second study was the CONKO1 clinical trial that investigated the role of adjuvant gemcitabine compared to observation. Tumor samples of 156 patients were analyzed for expression of hENT1. High hENT1 expression was not associated with improved median disease-free survival. In the gemcitabine group, disease-free survival was not significantly different at 13.2 months in low hENT1 patients vs. 11.5 months in high hENT1 patients (72). With these contradictory results and lack of prospective studies assessing the role of hENT1 in the adjuvant setting, nowadays hENT1 should not be used as a biomarker of response to gemcitabine.
CA19.9 in pancreatic cancer
CA19.9 is a Lewis blood group antigen, a carbohydrate produced by exocrine epithelial cells and is one of the most widely studied tumor markers in patients with pancreatic cancer (73). In patients with localized cancer, preoperative levels of CA19.9 cannot be used as a prognostic biomarker (74). Nevertheless, several studies have observed improved OS in patients with low CA19.9 levels after resection (75-77).
In the advanced pancreatic cancer setting, attempts have been made to evaluate the predictive value of basal CA19.9 levels and changes during treatment. In an analysis of six phase 2 trials of patients treated with gemcitabine-containing chemotherapy, changes in CA19.9 were predictive of outcome (78). In the phase 3 trial MPACT, in which the combination of gemcitabine plus nab-paclitaxel was evaluated and compared to gemcitabine alone, levels of CA19.9 and changes during treatment were evaluated. In a stepwise multivariate analysis, baseline CA 19.9 level was not an independent predictor of survival (79). Furthermore, an assessment to understand the dynamics of CA19.9 changes during treatment was pre-specified. CA19.9 was evaluated at baseline and every 8 weeks. Patients with CA19.9 decline (80%) versus patients without (20%) had improved OS (median 11.1 vs. 8.0 months, P=0.001) respectively. Almost all patients who achieved a partial or complete response in both arms had a decrease in CA19.9 levels. Among patients with stable disease, 79% of patients in the nab-paclitaxel arm and 78% in the gemcitabine alone arm had CA19.9 decrease at week 8, with corresponding median OS of 13.2 and 9.4 months respectively. Median OS for patients with stable disease but without CA 19.9 decrease at week 8 was 8.3 and 7.1 months respectively. In conclusion, CA 19.9 may be an early marker for antitumor activity in patients with metastatic pancreatic cancer and reduction in CA19.9 is an indicator of treatment benefit, particularly with the nab-paclitaxel regimen.
Secreted protein acidic and rich in cysteine (SPARC) in pancreatic cancer
SPARC has a role in tissue remodeling, embryonic development, cell migration and angiogenesis (80). The implication of SPARC in cancer is unclear, with most studies suggesting a pro-tumorogenic role, but some suggesting an anti-tumorigenic role (81). In pancreatic cancer, SPARC has been localized in tumor stroma, in fibroblasts and tumor epithelial cells. This protein has been evaluated as a prognostic and predictive biomarker.
In terms of its prognostic properties, two studies evaluated SPARC as a biomarker. The first, a retrospective analysis of 299 patients with resectable pancreatic cancer (82), SPARC positivity in stromal fibroblasts was associated with worse OS compared with SPARC negativity, with an HR of 1.89 (95% CI, 1.32–274; P=0.001). The presence of SPARC in tumor epithelial cells did not significantly correlate with OS. Similar results were observed in a sub-analysis of the CONKO-001 clinical trial. In this study both epithelia and fibroblast SPARC expression were associated with worse OS (83). Nevertheless, in this study, the effect of SPARC was restricted to patients treated with gemcitabine.
Furthermore, it was hypothesized that, taking in consideration the binding between SPARC and albumin, this could play a role in response to nab-paclitaxel (an albumin-based formulation of paclitaxel). In the phase 1–2 trial evaluating the safety and efficacy of nab-paclitaxel plus gemcitabine, higher SPARC expression was associated with longer OS (84). However, in the phase 3 MPACT study, SPARC levels were not associated with efficacy (80). In consequence, SPARC cannot be considered a prognostic or a predictive marker for response to gemcitabine and nab-paclitaxel.
Other biomarkers in pancreatic cancer
In the locally advanced setting, it is not clear if patients benefit from chemoradiotherapy. SMAD4 (Mothers against DPP Homolog 4) deletion is a promising biomarker that might identify patients who are less likely to benefit from locoregional strategies (85). The Radiation Therapy Oncology Group (RTOG) 1201 trial will stratify locally advanced pancreatic cancer patients according to SMAD4 status in order to evaluate this biomarker for prediction to response to therapy. Although this marker needs prospective validation, it appears to be a prognostic biomarker for selecting patients likely to develop metastatic disease.
Another interesting biomarker in pancreatic cancer is the loss of heterozygosity of STK11/LKB1, a tumor suppressor gene that encodes an inhibitor of mTOR, and is a characteristic of patients with Peutz-Jeghers Syndrome. A case report has shown benefit in a patient with this alteration who was treated with an mTOR inhibitor (86), indicating that SRK11 mutation could be a predictive biomarker.
Conclusions
Biomarkers are increasingly useful tools to predict prognosis and response to therapy in cancer patients. Furthermore, they allow us to improve our understanding of mechanisms of action and resistance to treatment. In this review we have evaluated the prognostic and predictive role of several biomarkers in colorectal, gastric and pancreatic cancer. In CRC, the value of RAS mutations has changed clinical practice facilitating a better outcome in a subgroup of patients, whilst avoiding unnecessary drug exposure in others. Furthermore, several biomarkers that will guide the development of novel therapeutic agents are emerging, potentially improving outcomes for some poor prognostic groups. In gastric cancer, the ToGA study is an example of a trial designed using selection by molecular marker, allowing the approval of anti-HER2 targeted agents in this histology. In pancreatic cancer patients, although there is a need to validate various research axes investigated to date, the BRCA mutation is a predictive biomarker to response to platinum that will improve outcomes in these patients and may also be used to direct the development of PARP inhibitors. Furthermore, CA19.9 can be exploited to assess prognosis and response to therapy.
To ensure that new discoveries reach the clinic as rapidly as possible, the evaluation of biomarkers is a critical aspect to integrate when designing clinical trials. Moreover, selecting patients by molecular markers will benefit drug development, permitting earlier approvals as per unselected population. In support of this, increased efforts are needed to validate biomarkers, and preclinical and clinical studies are needed to identify biomarkers that will improve our patients’ chances of increased survival by offering them the optimal therapies and limiting toxicity.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- GLOBOCAN 2008: Cancer Incidence and Mortality Worldwide. International Agency for Research on Cancer, 2010. Available online: https://www.iarc.fr/en/media-centre/iarcnews/2010/globocan2008.php
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 2003;3:11-22. [Crossref] [PubMed]
- Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005;5:341-54. [Crossref] [PubMed]
- Andreyev HJ, Norman A, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’study. Br J Cancer 2001;85:692. [Crossref] [PubMed]
- Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369:1023-34. [Crossref] [PubMed]
- Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 2004;351:337-45. [Crossref] [PubMed]
- Van Cutsem E, Peeters M, Siena S, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol 2007;25:1658-64. [Crossref] [PubMed]
- Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:1626-34. [Crossref] [PubMed]
- Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008;359:1757-65. [Crossref] [PubMed]
- Van Cutsem E, Lenz H-J, Köhne C-H, et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J Clin Oncol 2015;33:692-700. [Crossref] [PubMed]
- Jonker DJ, O'Callaghan CJ, Karapetis CS, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med 2007;357:2040-8. [Crossref] [PubMed]
- Douillard JY, Siena S, Cassidy J, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol 2010;28:4697-705. [Crossref] [PubMed]
- Bokemeyer C, Bondarenko I, Hartmann J, et al. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol 2011;22:1535-46. [Crossref] [PubMed]
- Benson AB 3rd, Bekaii-Saab T, Chan E, et al. Metastatic colon cancer, version 3.2013: featured updates to the NCCN Guidelines. J Natl Compr Canc Netw 2013;11:141-52. [Crossref] [PubMed]
- Loupakis F, Ruzzo A, Cremolini C, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer 2009;101:715-21. [Crossref] [PubMed]
- Peeters M, Oliner KS, Parker A, et al. Massively parallel tumor multigene sequencing to evaluate response to panitumumab in a randomized phase III study of metastatic colorectal cancer. Clin Cancer Res 2013;19:1902-12. [Crossref] [PubMed]
- De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010;11:753-62. [Crossref] [PubMed]
- Lenz HJ, Lee FC, Yau L, et al. MAVERICC, a phase 2 study of mFOLFOX6-bevacizumab (BV) vs FOLFIRI-BV with biomarker stratification as first-line (1L) chemotherapy (CT) in patients (pts) with metastatic colorectal cancer (mCRC). J Clin Oncol 2016;34:abstr 493.
- Tabernero J, Lenz H-J, Siena S, et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: a retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol 2015;16:937-48. [Crossref] [PubMed]
- Tejpar S, Bertagnolli M, Bosman F, et al. Prognostic and predictive biomarkers in resected colon cancer: current status and future perspectives for integrating genomics into biomarker discovery. Oncologist 2010;15:390-404. [Crossref] [PubMed]
- Lee EJ, Choi C, Park CK, et al. Tracing origin of serrated adenomas with BRAF and KRAS mutations. Virchows Arch 2005;447:597-602. [Crossref] [PubMed]
- Chan TL, Zhao W, Leung SY, et al. BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res 2003;63:4878-81. [PubMed]
- Rajagopalan H, Bardelli A, Lengauer C, et al. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature 2002;418:934. [Crossref] [PubMed]
- Fransén K, Klintenäs M, Österström A, et al. Mutation analysis of the BRAF, ARAF and RAF-1 genes in human colorectal adenocarcinomas. Carcinogenesis 2004;25:527-33. [Crossref] [PubMed]
- Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res 2005;65:6063-9. [Crossref] [PubMed]
- Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 2008;26:5705-12. [Crossref] [PubMed]
- Corcoran RB, Atreya CE, Falchook GS, et al. Combined BRAF and MEK inhibition with dabrafenib and trametinib in BRAF V600–mutant colorectal cancer. J Clin Oncol 2015;33:4023-31. [Crossref] [PubMed]
- Atreya CE, Van Cutsem E, Bendell JC, et al. Updated efficacy of the MEK inhibitor trametinib (T), BRAF inhibitor dabrafenib (D), and anti-EGFR antibody panitumumab (P) in patients (pts) with BRAF V600E mutated (BRAFm) metastatic colorectal cancer (mCRC). J Clin Oncol 2015;33:abstr 103.
- Tabernero J, Van Geel R, Guren TK, et al. Phase 2 results: Encorafenib (ENCO) and cetuximab (CETUX) with or without alpelisib (ALP) in patients with advanced BRAF-mutant colorectal cancer (BRAFm CRC). J Clin Oncol 2016;34:abstr 3544.
- de la Chapelle A, Hampel H. Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol 2010;28:3380-7. [Crossref] [PubMed]
- Popat S, Hubner R, Houlston R. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 2005;23:609-18. [Crossref] [PubMed]
- Guastadisegni C, Colafranceschi M, Ottini L, et al. Microsatellite instability as a marker of prognosis and response to therapy: a meta-analysis of colorectal cancer survival data. Eur J Cancer 2010;46:2788-98. [Crossref] [PubMed]
- Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med 2003;349:247-57. [Crossref] [PubMed]
- Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol 2010;28:3219-26. [Crossref] [PubMed]
- Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015;5:43-51. [Crossref] [PubMed]
- Smyrk TC, Watson P, Kaul K, et al. Tumor-infiltrating lymphocytes are a marker for microsatellite instability in colorectal carcinoma. Cancer 2001;91:2417-22. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Tol J, Dijkstra JR, Klomp M, et al. Markers for EGFR pathway activation as predictor of outcome in metastatic colorectal cancer patients treated with or without cetuximab. Eur J Cancer 2010;46:1997-2009. [Crossref] [PubMed]
- Barbara C, Martin V, Molinari F, et al. Use of HER2 gene amplification to identify patients with metastatic colorectal cancer resistant to anti-EGFR monoclonal antibodies. J Clin Oncol 2012;30:abstr 474.
- Sartore-Bianchi A, Trusolino L, Martino C, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol 2016;17:738-46. [Crossref] [PubMed]
- Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350-6. [Crossref] [PubMed]
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol 2006;24:2137-50. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Ménard S, Pupa SM, Campiglio M, et al. Biologic and therapeutic role of HER2 in cancer. Oncogene 2003;22:6570-8. [Crossref] [PubMed]
- Gravalos C, Jimeno A. HER2 in gastric cancer: a new prognostic factor and a novel therapeutic target. Ann Oncol 2008;19:1523-9. [Crossref] [PubMed]
- Jørgensen JT, Hersom M. HER2 as a prognostic marker in gastric cancer-a systematic analysis of data from the literature. J Cancer 2012;3:137-44. [Crossref] [PubMed]
- Gu J, Zheng L, Wang Y, et al. Prognostic significance of HER2 expression based on trastuzumab for gastric cancer (ToGA) criteria in gastric cancer: an updated meta-analysis. Tumour Biol 2014;35:5315-21. [Crossref] [PubMed]
- Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687-97. [Crossref] [PubMed]
- Alsina M, Moehler M, Hierro C, et al. Immunotherapy for Gastric Cancer: A Focus on Immune Checkpoints. Target Oncol 2016;11:469-77. [Crossref] [PubMed]
- Bang YJ, Chung HC, Shankaran V, et al. Relationship between PD-L1 expression and clinical outcomes in patients with advanced gastric cancer treated with the anti-PD-1 monoclonal antibody pembrolizumab (MK-3475) in KEYNOTE-012. J Clin Oncol 2015;33:abstr 4001.
- Bang YJ, Chung HC, Shankaran V, et al. Clinical outcomes and their correlation with gene expression in patients with advanced gastric cancer treated with pembrolizumab (MK-3475): KEYNOTE-012. Ann Oncol 2015;26:iv118. [Crossref]
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin 2009;59:225-49. [Crossref] [PubMed]
- Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012;491:399-405. [Crossref] [PubMed]
- Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495-501. [Crossref] [PubMed]
- Shah MA, Kurtz RC. Upper gastrointestinal cancer predisposition syndromes. Hematol Oncol Clin North Am 2010;24:815-35. [Crossref] [PubMed]
- Iqbal J, Ragone A, Lubinski J, et al. The incidence of pancreatic cancer in BRCA1 and BRCA2 mutation carriers. Br J Cancer 2012;107:2005-9. [Crossref] [PubMed]
- Couch FJ, Johnson MR, Rabe KG, et al. The prevalence of BRCA2 mutations in familial pancreatic cancer. Cancer Epidemiol Biomarkers Prev 2007;16:342-6. [Crossref] [PubMed]
- Ferrone CR, Levine DA, Tang LH, et al. BRCA germline mutations in Jewish patients with pancreatic adenocarcinoma. J Clin Oncol 2009;27:433-8. [Crossref] [PubMed]
- Lucas AL, Shakya R, Lipsyc MD, et al. High prevalence of BRCA1 and BRCA2 germline mutations with loss of heterozygosity in a series of resected pancreatic adenocarcinoma and other neoplastic lesions. Clin Cancer Res 2013;19:3396-403. [Crossref] [PubMed]
- Goggins M, Schutte M, Lu J, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res 1996;56:5360-4. [PubMed]
- Holter S, Borgida A, Dodd A, et al. Germline BRCA Mutations in a Large Clinic-Based Cohort of Patients With Pancreatic Adenocarcinoma. J Clin Oncol 2015;33:3124-9. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Golan T, Kanji Z, Epelbaum R, et al. Overall survival and clinical characteristics of pancreatic cancer in BRCA mutation carriers. Br J Cancer 2014;111:1132-8. [Crossref] [PubMed]
- Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015;33:244-50. [Crossref] [PubMed]
- Mackey JR, Mani RS, Selner M, et al. Functional nucleoside transporters are required for gemcitabine influx and manifestation of toxicity in cancer cell lines. Cancer Res 1998;58:4349-57. [PubMed]
- Spratlin J, Sangha R, Glubrecht D, et al. The absence of human equilibrative nucleoside transporter 1 is associated with reduced survival in patients with gemcitabine-treated pancreas adenocarcinoma. Clin Cancer Res 2004;10:6956-61. [Crossref] [PubMed]
- Giovannetti E, Del Tacca M, Mey V, et al. Transcription analysis of human equilibrative nucleoside transporter-1 predicts survival in pancreas cancer patients treated with gemcitabine. Cancer Res 2006;66:3928-35. [Crossref] [PubMed]
- Maréchal R, Bachet JB, Mackey JR, et al. Levels of gemcitabine transport and metabolism proteins predict survival times of patients treated with gemcitabine for pancreatic adenocarcinoma. Gastroenterology 2012;143:664-74.e1-6.
- Murata Y, Hamada T, Kishiwada M, et al. Human equilibrative nucleoside transporter 1 expression is a strong independent prognostic factor in UICC T3—T4 pancreatic cancer patients treated with preoperative gemcitabine-based chemoradiotherapy. J Hepatobiliary Pancreat Sci 2012;19:413-25. [Crossref] [PubMed]
- Greenhalf W, Ghaneh P, Neoptolemos JP, et al. Pancreatic cancer hENT1 expression and survival from gemcitabine in patients from the ESPAC-3 trial. J Natl Cancer Inst 2014;106:djt347. [Crossref] [PubMed]
- Sinn M, Riess H, Sinn BV, et al. Human equilibrative nucleoside transporter 1 expression analysed by the clone SP 120 rabbit antibody is not predictive in patients with pancreatic cancer treated with adjuvant gemcitabine - Results from the CONKO-001 trial. Eur J Cancer 2015;51:1546-54. [Crossref] [PubMed]
- Narimatsu H, Iwasaki H, Nakayama F, et al. Lewis and secretor gene dosages affect CA19-9 and DU-PAN-2 serum levels in normal individuals and colorectal cancer patients. Cancer Res 1998;58:512-8. [PubMed]
- Winter JM, Yeo CJ, Brody JR. Diagnostic, prognostic, and predictive biomarkers in pancreatic cancer. J Surg Oncol 2013;107:15-22. [Crossref] [PubMed]
- Glenn J, Steinberg WM, Kurtzman SH, et al. Evaluation of the utility of a radioimmunoassay for serum CA 19-9 levels in patients before and after treatment of carcinoma of the pancreas. J Clin Oncol 1988;6:462-8. [Crossref] [PubMed]
- Berger AC, Garcia M, Hoffman JP, et al. Postresection CA 19-9 predicts overall survival in patients with pancreatic cancer treated with adjuvant chemoradiation: a prospective validation by RTOG 9704. J Clin Oncol 2008;26:5918-22. [Crossref] [PubMed]
- Kinsella TJ, Seo Y, Willis J, et al. The impact of resection margin status and postoperative CA19-9 levels on survival and patterns of recurrence after postoperative high-dose radiotherapy with 5-FU-based concurrent chemotherapy for resectable pancreatic cancer. Am J Clin Oncol 2008;31:446-53. [Crossref] [PubMed]
- Bauer TM, El-Rayes BF, Li X, et al. Carbohydrate antigen 19-9 is a prognostic and predictive biomarker in patients with advanced pancreatic cancer who receive gemcitabine-containing chemotherapy. Cancer 2013;119:285-92. [Crossref] [PubMed]
- Tabernero J, Chiorean EG, Infante JR, et al. Prognostic factors of survival in a randomized phase III trial (MPACT) of weekly nab-paclitaxel plus gemcitabine versus gemcitabine alone in patients with metastatic pancreatic cancer. Oncologist 2015;20:143-50. [Crossref] [PubMed]
- Hidalgo M, Plaza C, Musteanu M, et al. SPARC Expression Did Not Predict Efficacy of nab-Paclitaxel plus Gemcitabine or Gemcitabine Alone for Metastatic Pancreatic Cancer in an Exploratory Analysis of the Phase III MPACT Trial. Clin Cancer Res 2015;21:4811-8. [Crossref] [PubMed]
- Podhajcer OL, Benedetti LG, Girotti MR, et al. The role of the matricellular protein SPARC in the dynamic interaction between the tumor and the host. Cancer Metastasis Rev 2008;27:691-705. [Crossref] [PubMed]
- Infante JR, Matsubayashi H, Sato N, et al. Peritumoral fibroblast SPARC expression and patient outcome with resectable pancreatic adenocarcinoma. J Clin Oncol 2007;25:319-25. [Crossref] [PubMed]
- Sinn M, Sinn B, Striefler J, et al. SPARC expression in resected pancreatic cancer patients treated with gemcitabine: results from the CONKO-001 study. Ann Oncol 2014;25:1025-32. [Crossref] [PubMed]
- Von Hoff DD, Ramanathan RK, Borad MJ, et al. Gemcitabine plus nab-paclitaxel is an active regimen in patients with advanced pancreatic cancer: a phase I/II trial. J Clin Oncol 2011;29:4548-54. [Crossref] [PubMed]
- Iacobuzio-Donahue CA, Fu B, Yachida S, et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol 2009;27:1806-13. [Crossref] [PubMed]
- Klümpen HJ, Queiroz KC, Spek CA, et al. mTOR inhibitor treatment of pancreatic cancer in a patient With Peutz-Jeghers syndrome. J Clin Oncol 2011;29:e150-3. [Crossref] [PubMed]


